
|
|
|
The anaphase-promoting complex is an E3 ligase that is used to determine the
progression of mitosis by decorating specific molecules with ubiquitin in a
programmed manner. It has 16 named subunits and functions as the counter to
transcription for mitosis-related proteins, e.g., securin and
cyclin B.
α⧸ω ST phosphorylation diagrams for human & mouse anaphase-promoting complex subunit 1 (#ANAPC1:p). The human & mouse patterns have an unusual degree of similarity, with phospho-IDR sprouting up between structured regions in the N-terminal third of the sequence. All but the most nward are S/TP-type acceptors. #proteomics #anapcs 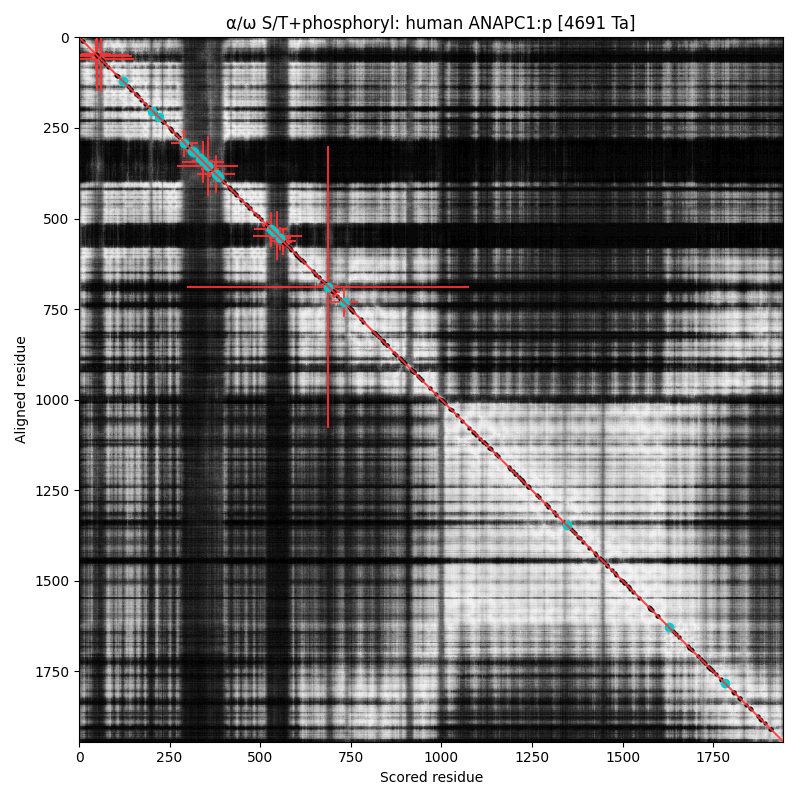 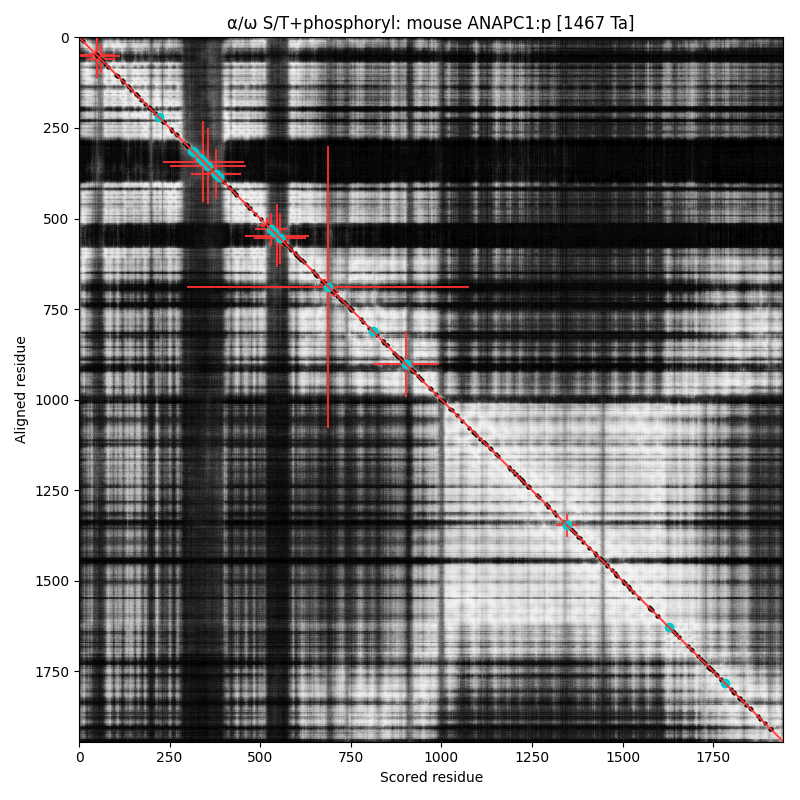
α⧸ω ST phosphorylation diagrams for human & mouse anaphase-promoting complex subunit 2 (#ANAPC2:p). The human & mouse patterns are very similar, with the catalytic cullin domain (479-718) bracketed by an nward high occupancy phospho-IDR and a cward low occupancy pIDR. #proteomics #anapcs 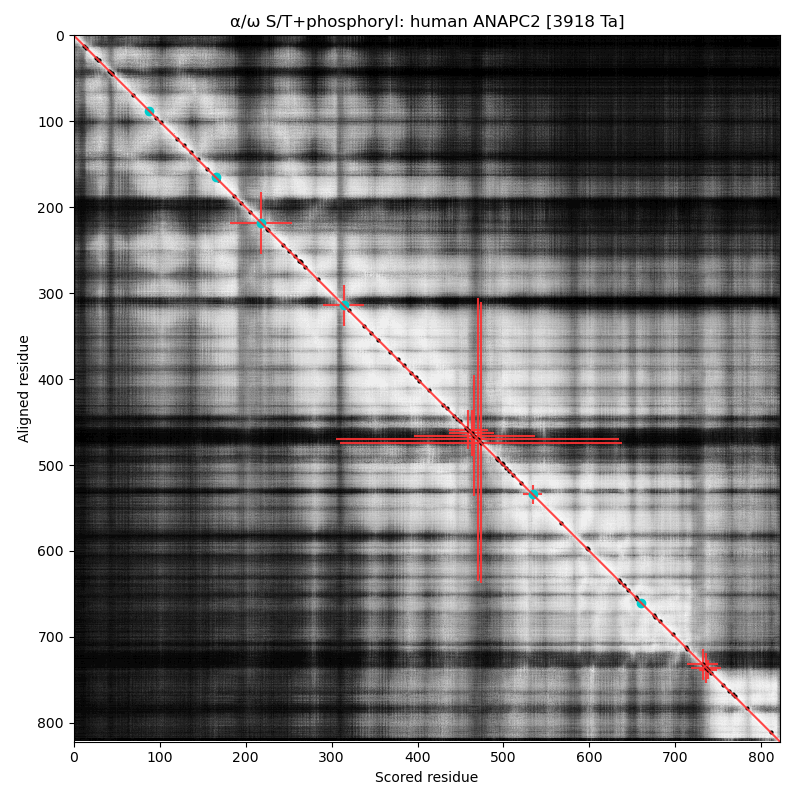 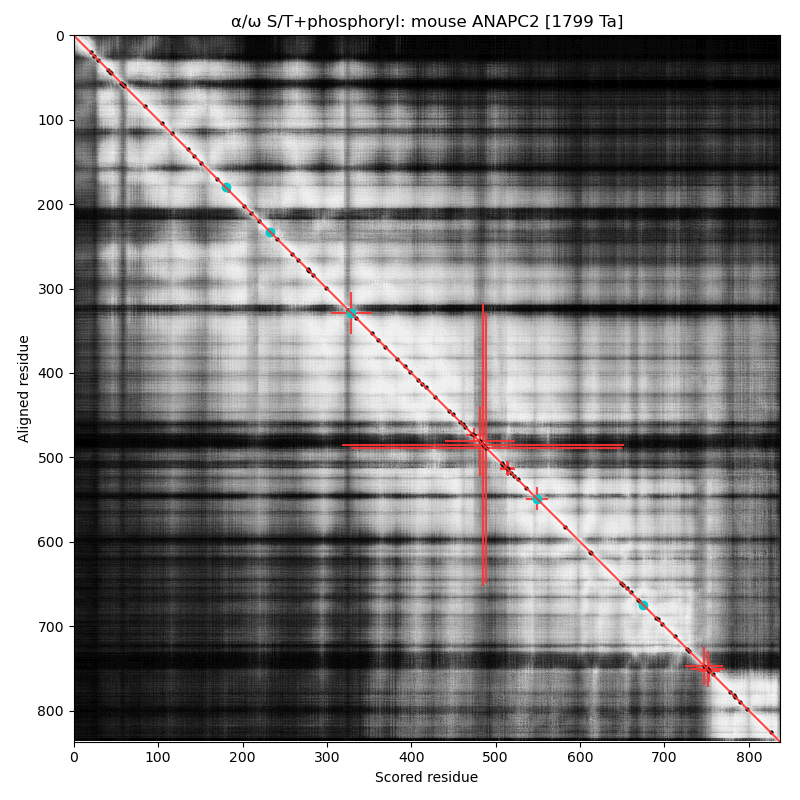
α⧸ω ST phosphorylation diagrams for human & mouse cell division cycle 27 (#CDC27, #ANAPC3:p). The human & mouse patterns share a common phospho-IDR, situated between nward & cward tetratricopeptide-like helical domains. Several subunits in the anaphase-promoting complex have older "cell division cycle" names that are still being used. #proteomics #anapc 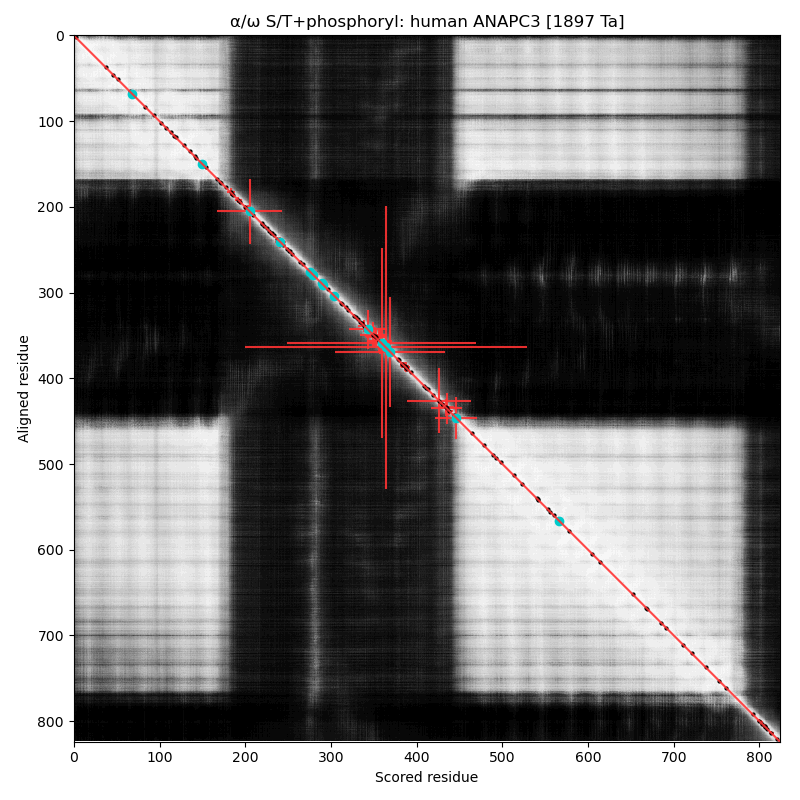 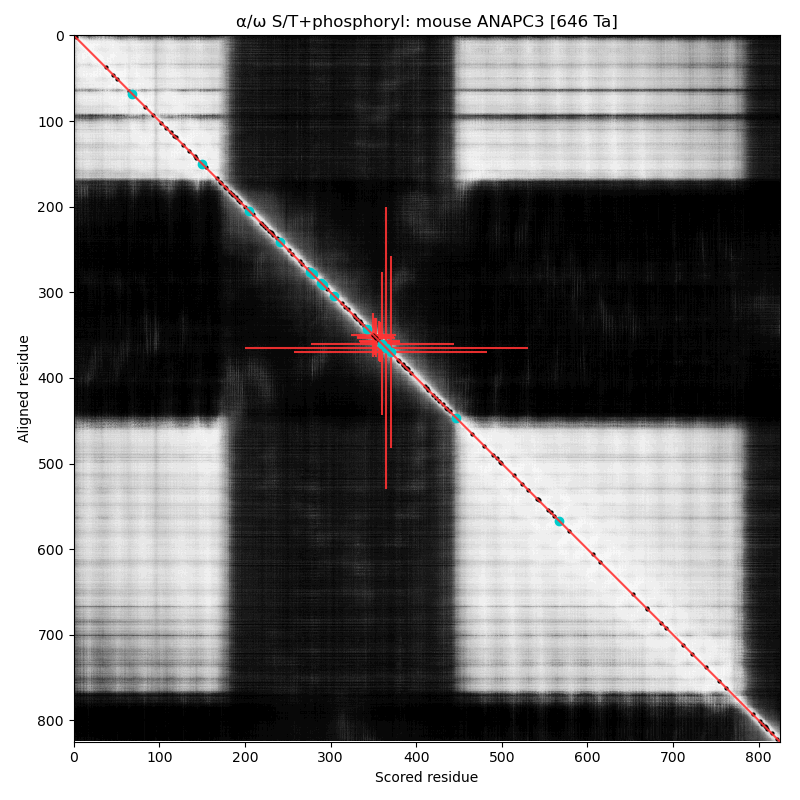
α⧸ω ST phosphorylation diagrams for human & mouse anaphase promoting complex subunit 4 (#ANAPC4:p). They share a common C-terminal dragon's tail phospho-IDR, with observed multiple occupancy. The structured domains are typical for protein-protein interactions: an nward, all-strands WD40 repeat and a 4 helix coiled-coil. #proteomics #anapc 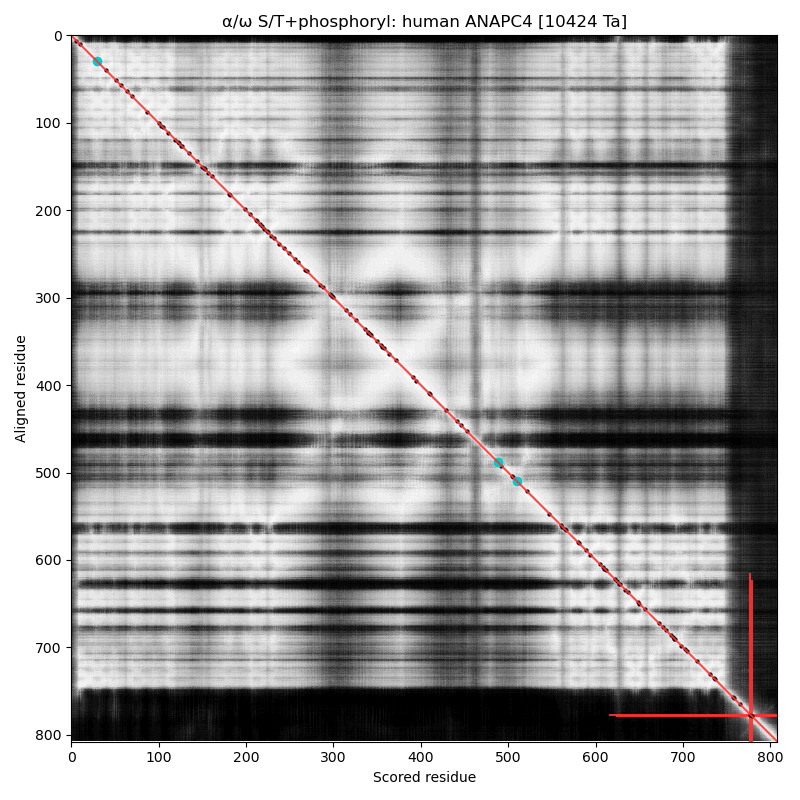 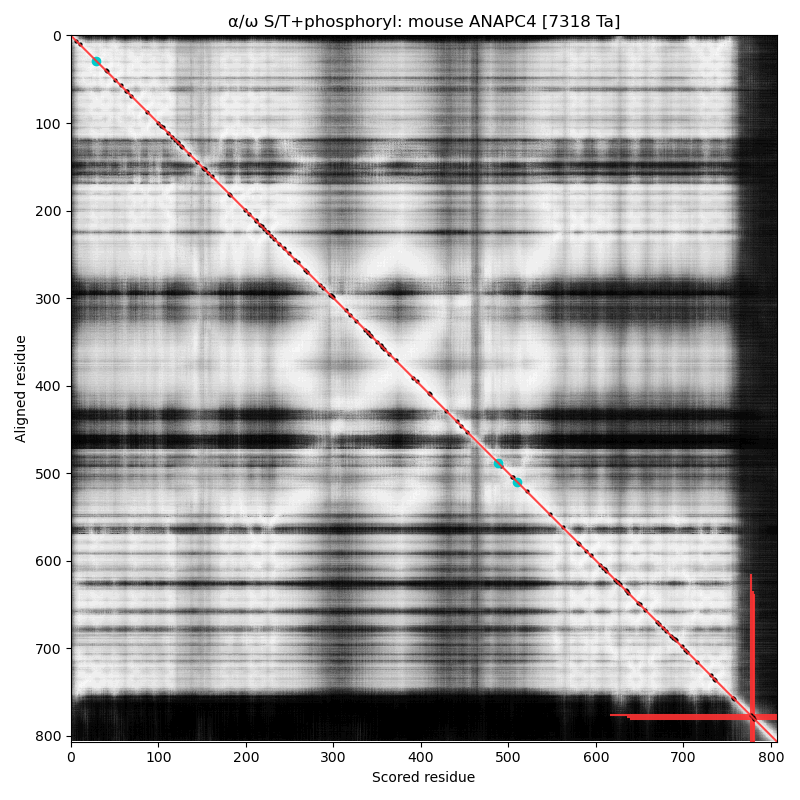
α⧸ω ST phosphorylation diagrams for human & mouse anaphase promoting complex subunit 5 (#ANAPC5:p). The human sequence has several low occupancy acceptors sprout up in short phosphoIDRs that connect helix-rich domains that take up most of the real estate. #proteomics #anapc 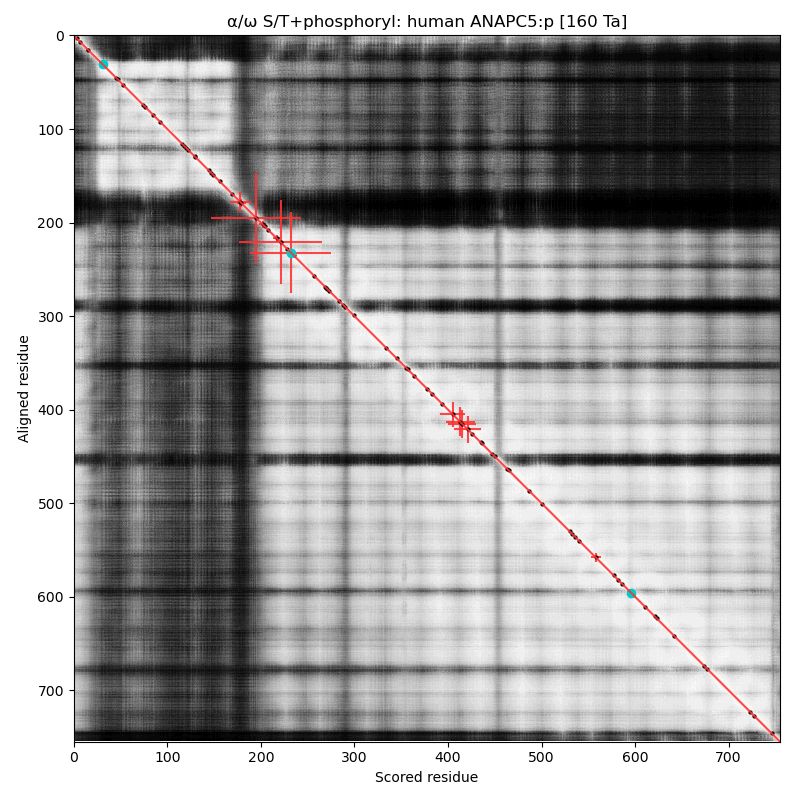 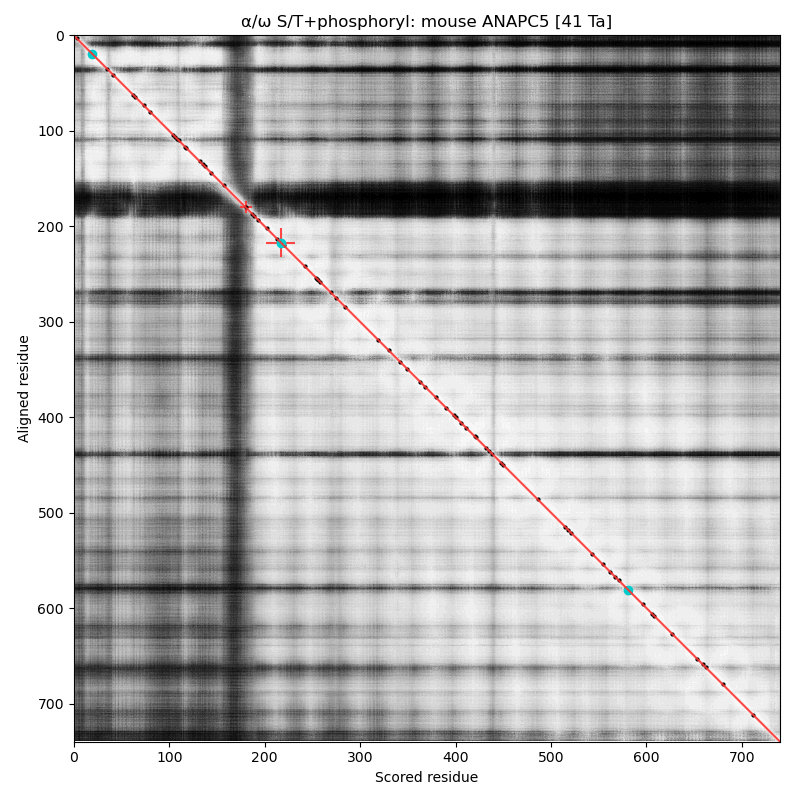
α⧸ω ST phosphorylation diagrams for human & mouse cell division cycle 16 (#CDC16, aka #ANAPC6:p). The sequences have a C-terminal dragon's tail phosphoIDRs with 2 S/TP type acceptors. The structured nward domain is a cluster of tetratricopeptide repeat helix bundles. #proteomics #anapc 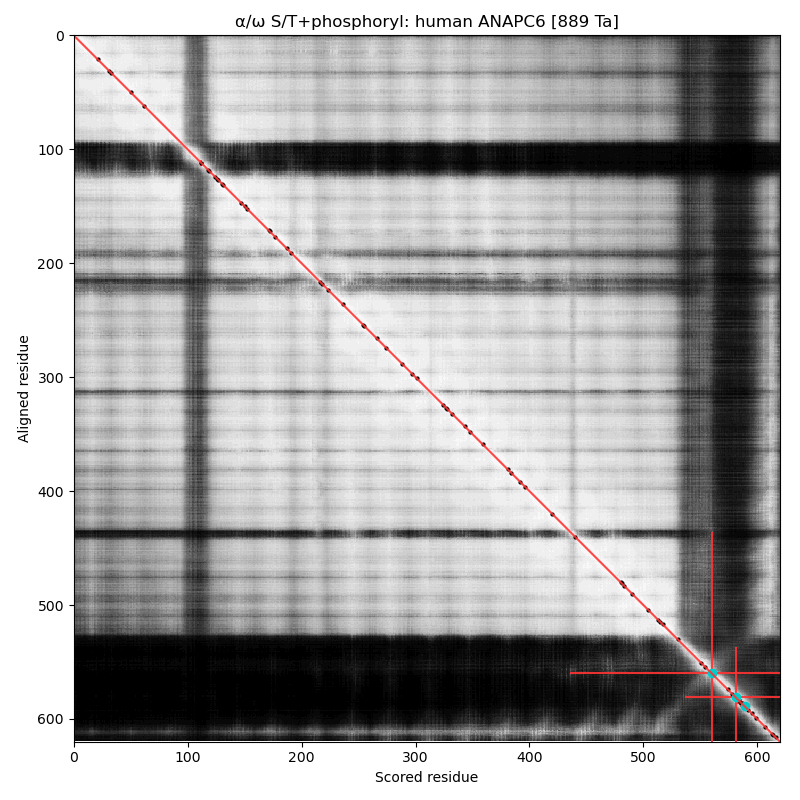 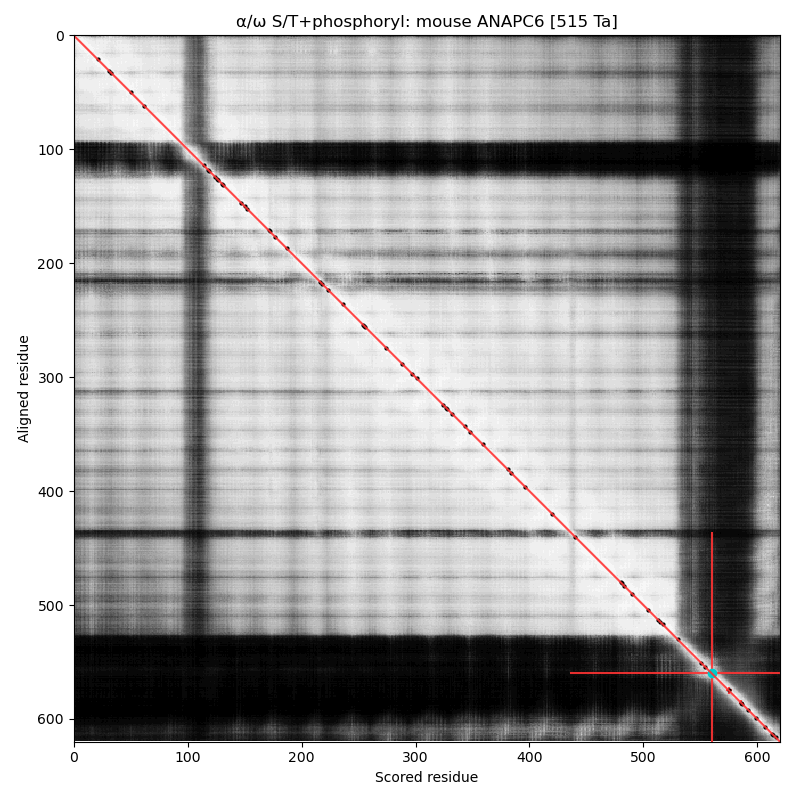
α⧸ω ST phosphorylation diagrams for human & mouse anaphase promoting complex subunit 7 (#ANAPC7:p). The human sequence has 2 low occupancy phosphoIDRs adjacent to the tetratricopeptide repeat helix bundles that dominate the shiny parts. #proteomics #anapc  
α⧸ω ST phosphorylation diagrams for human & mouse cell division cycle 23 (#CDC23:p, aka #ANAPC8:p). Both sequences have multiple high occupancy S/TP-type acceptors on a short dragon's tail phosphoIDR. The nward bulk of the protein is composed of phosphorylation-resistant tetratricopeptide repeats. #proteomics #anapc  
α⧸ω ST phosphorylation diagrams for yeast anaphase promoting complex subunit 9 (#APC9:p). This subunit is present in yeast but there is no homolog in the mammalian complex. It has a single high occupancy receptor, T36+phosphoryl. #proteomics #anapc 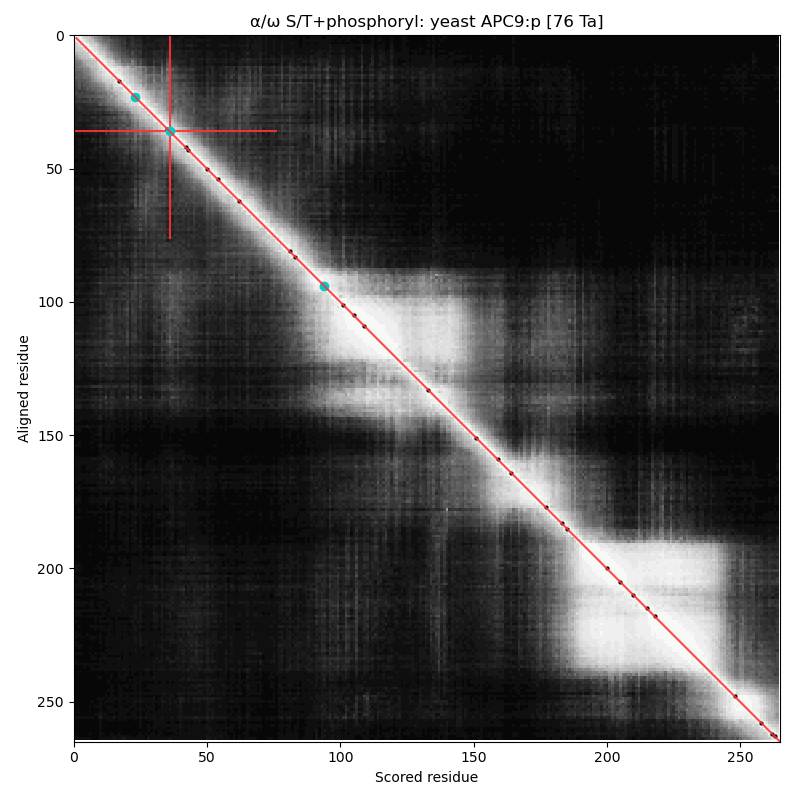
α⧸ω ST phosphorylation diagrams for human & mouse anaphase promoting complex subunit 10 (#ANAPC10:p). This subunit does not have any high occupancy phosphorylation acceptors, although both sequence have a low occupancy acceptor T7+phosphoryl. #proteomics #anapc 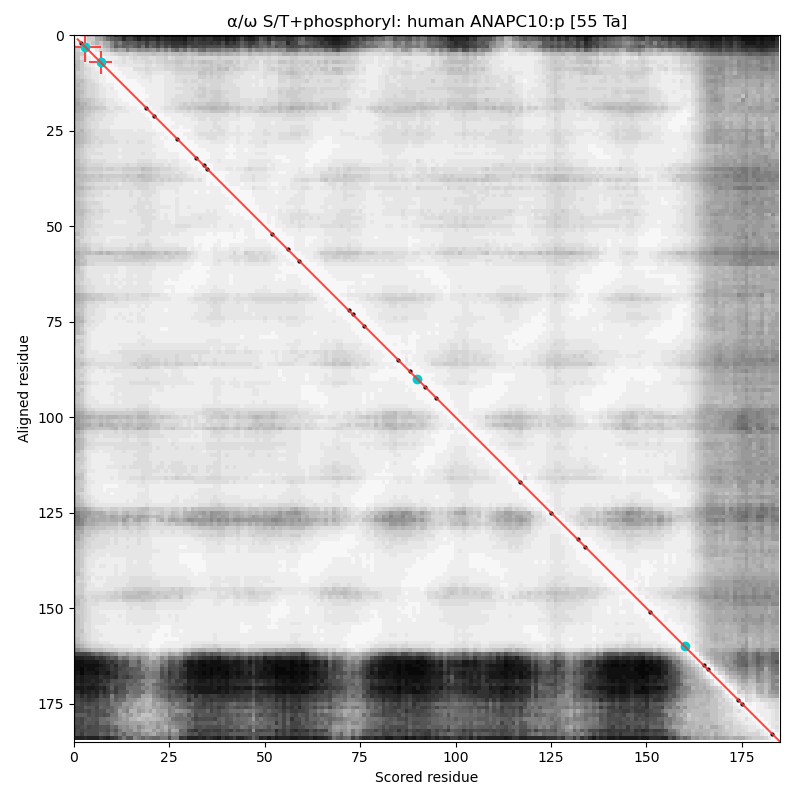 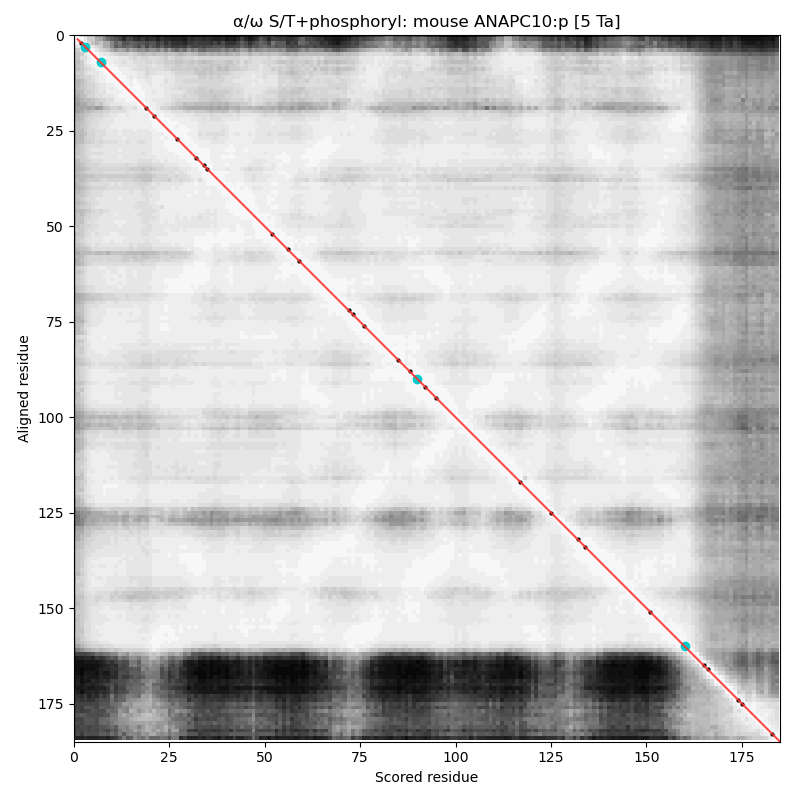
α⧸ω ST phosphorylation diagrams for human & mouse cell divlsion cycle 20 (#CDC20:p). Both sequences have a dragon's neck type N-terminal phosphoIDR with three high occupancy S/TP acceptors. Phosphorylation is known to prevent association with the anaphase promoting complex. The cward shiny region is a kinase-resistant, WD40 repeat beta-propeller. #proteomics #anapc 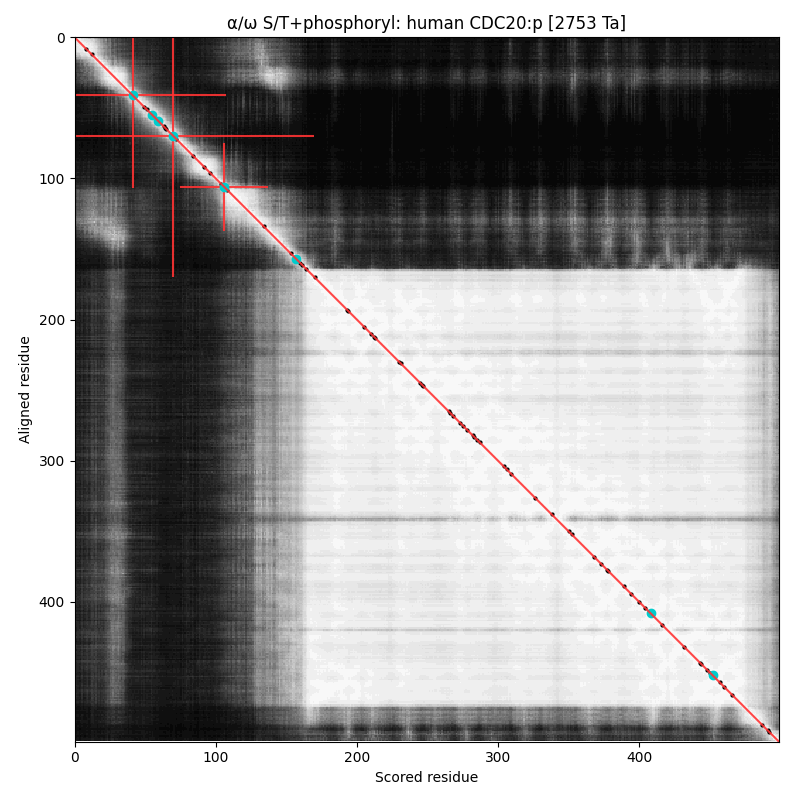 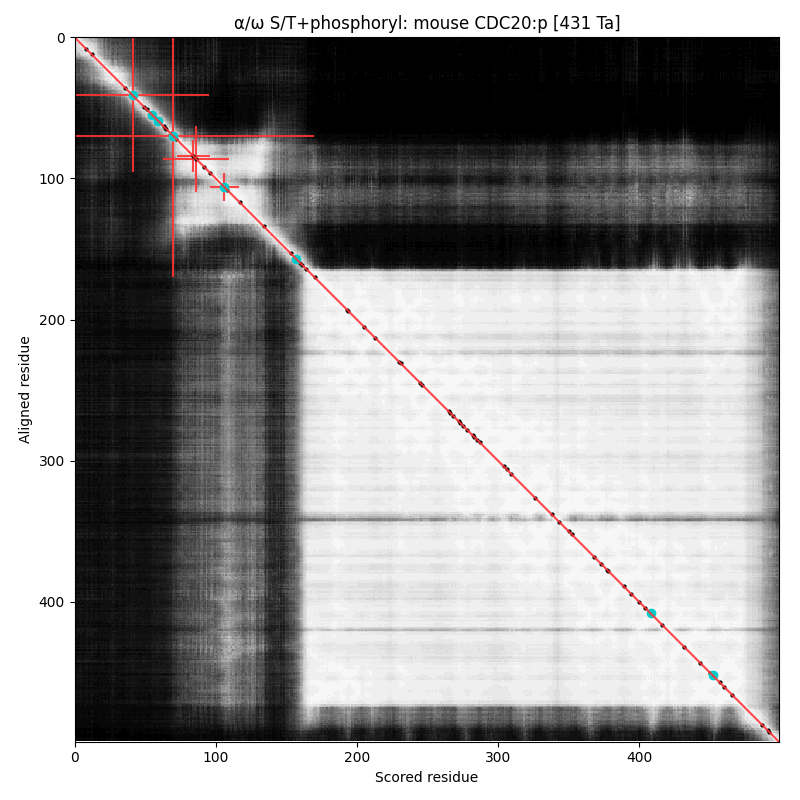
Anaphase-promoting complex has a discrete set of substrates that allow
it to control the pace of mitosis.
A GPMDB ubiquitination PTM diagram for human Pituitary Tumour Transforming Gene 1 regulator of sister chromatid separation, securin (PTTG1:p). This protein is one of the substrates of the anaphase-promoting complex, a very special E3 ubiquitin ligase. It is part of the mitosis coördination program that runs in all cells during M phase & is in no way limited to tumours. 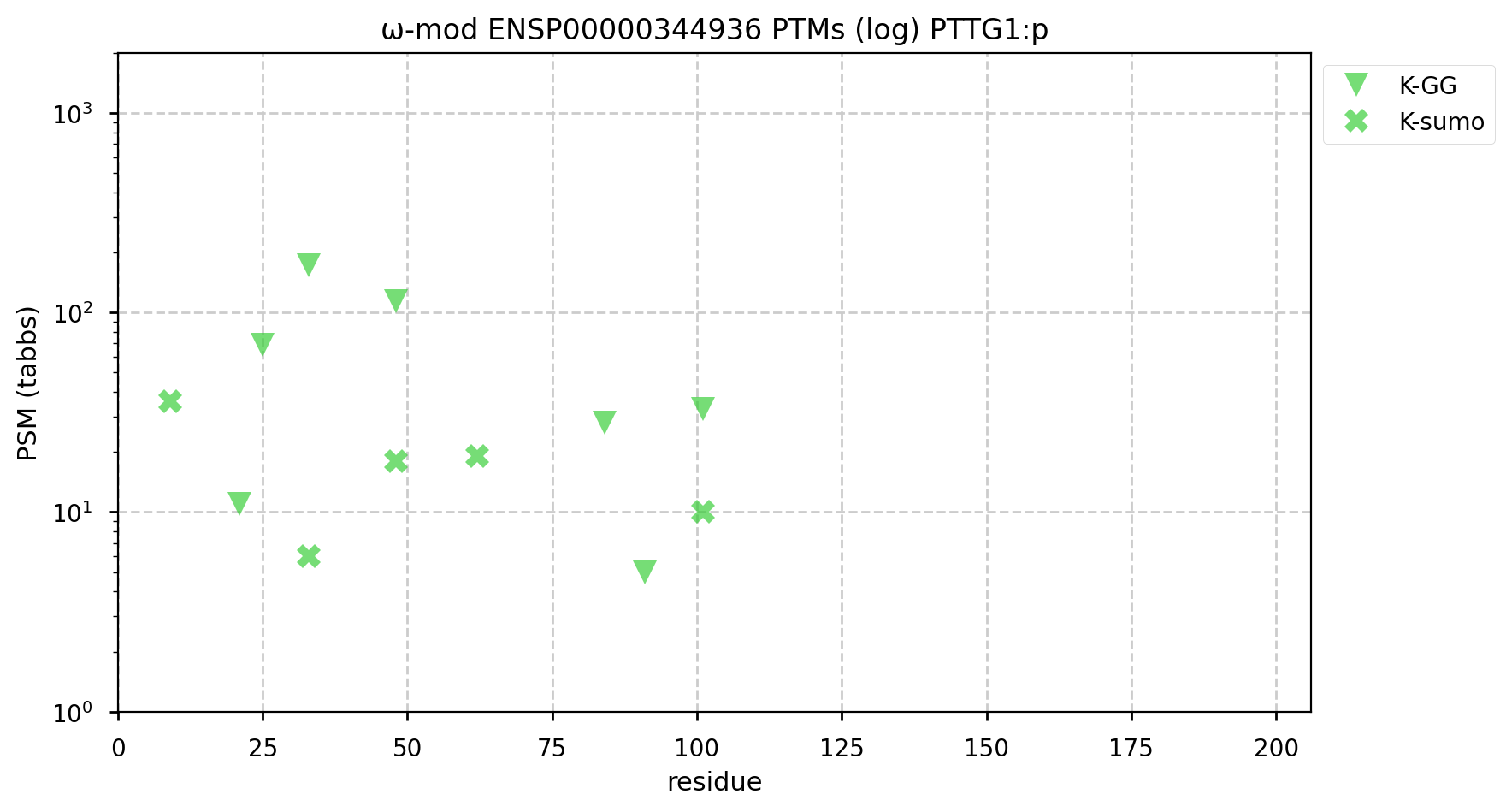
A GPMDB ubiquitination PTM diagram for human cyclin B1 (CCNB1:p). This protein is one of the substrates of the anaphase-promoting complex. This cell cycle promoter is activated by phosphorylation during prophase & must be removed via ubiquitination for anaphase to proceed. https://www.nature.com/scitable/topicpage/mitosis-and-cell-division-205/ 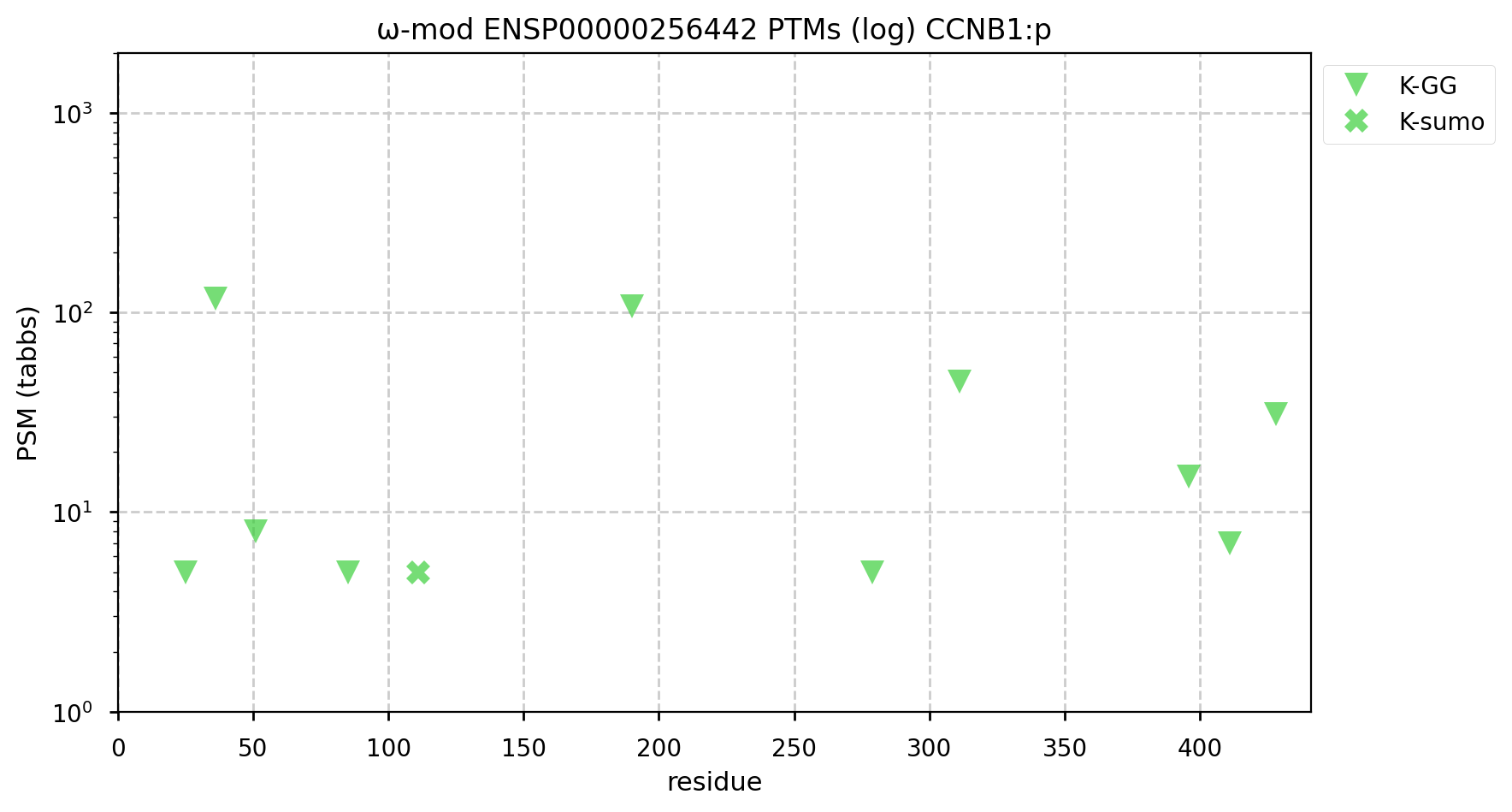
A GPMDB ubiquitination PTM diagram for human SCL/TAL1 interrupting locus centriolar assembly protein (STIL:p). There is evidence that it is a ubiquitination target of the anaphase-promoting complex (https://pubmed.ncbi.nlm.nih.gov/22349698), resulting in a rapid decline in concentration at the metaphase-to-anaphase transition. #proteomics #anapc 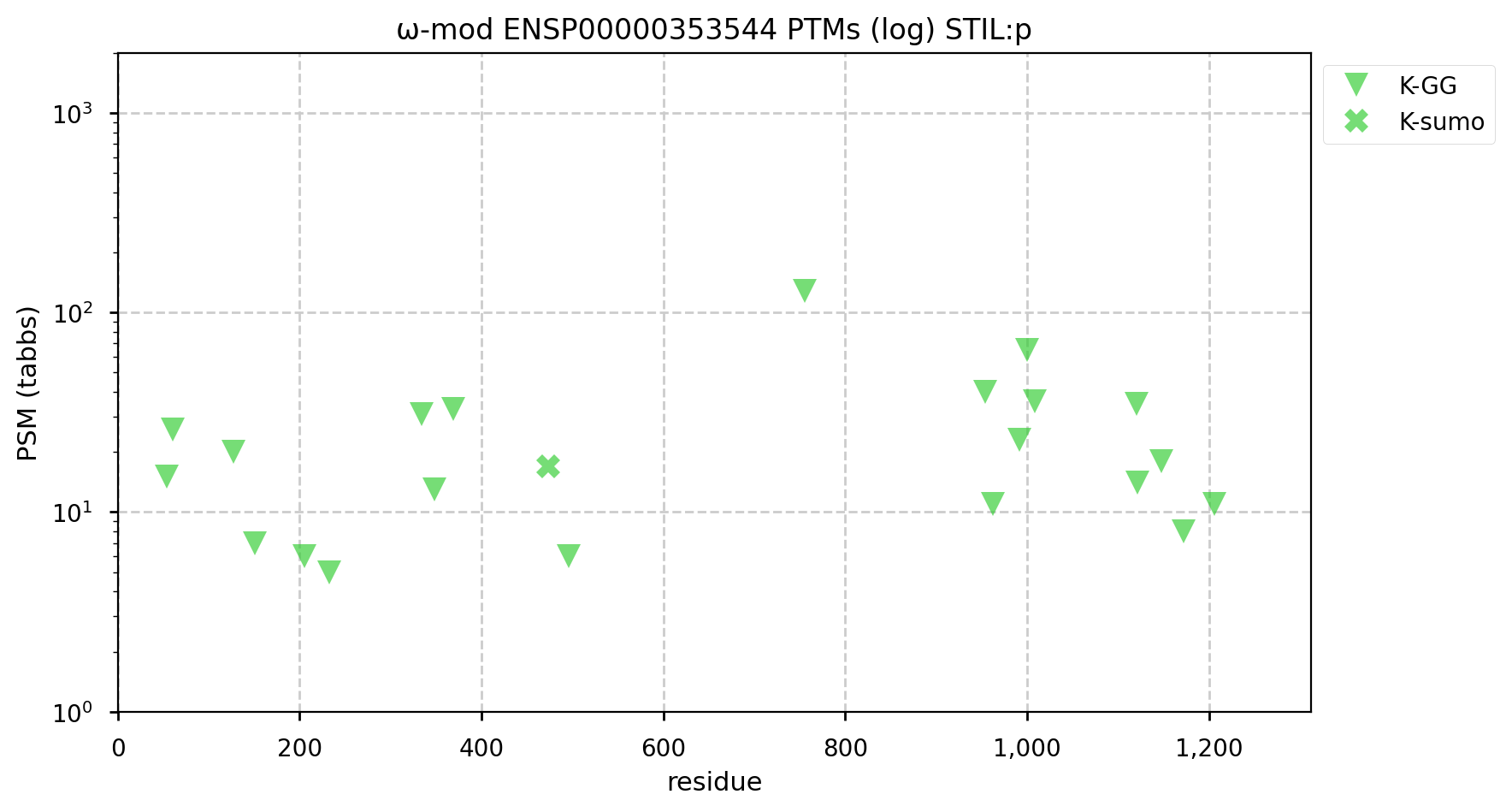
A GPMDB ubiquitination PTM diagram for human BTG3 associated nuclear protein (#BANP:p). There is evidence that it is a ubiquitination target of the anaphase-promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5520925/). Some of the observed K+Ub acceptors may also be occupied by K+SUMO2/3. #proteomics #anapc 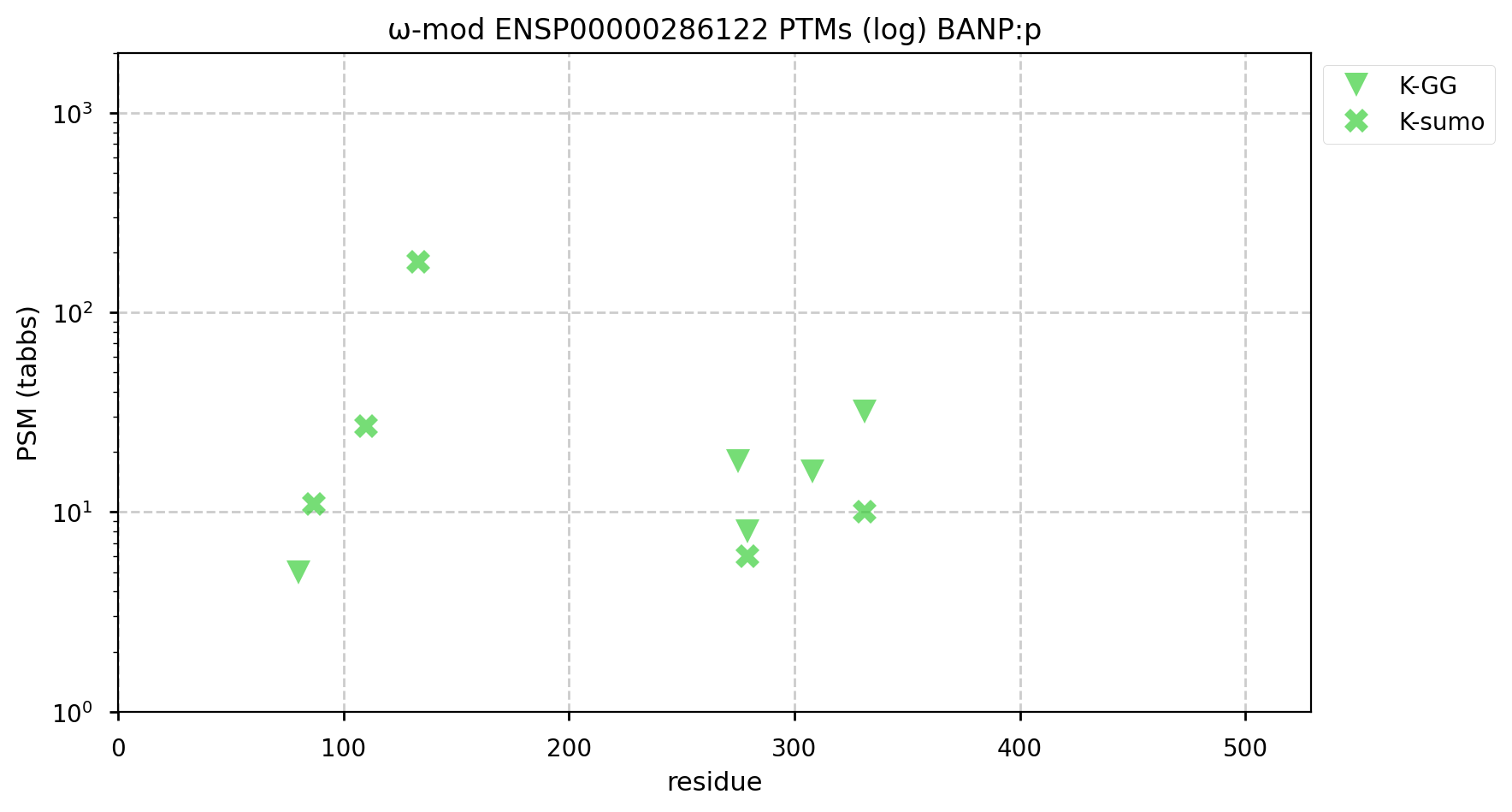
A GPMDB ubiquitination PTM diagram for human never-in-mitosis gene A―NIMA―related kinase 2 (#NEK2:p). The 'zines suggest that it is a ubiquitination target of the anaphase-promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6927175/). While there are some low occupancy K+SUMO2/3 modifications, the heavy lifting seems to be left up to K+Ub. #proteomics #anapc 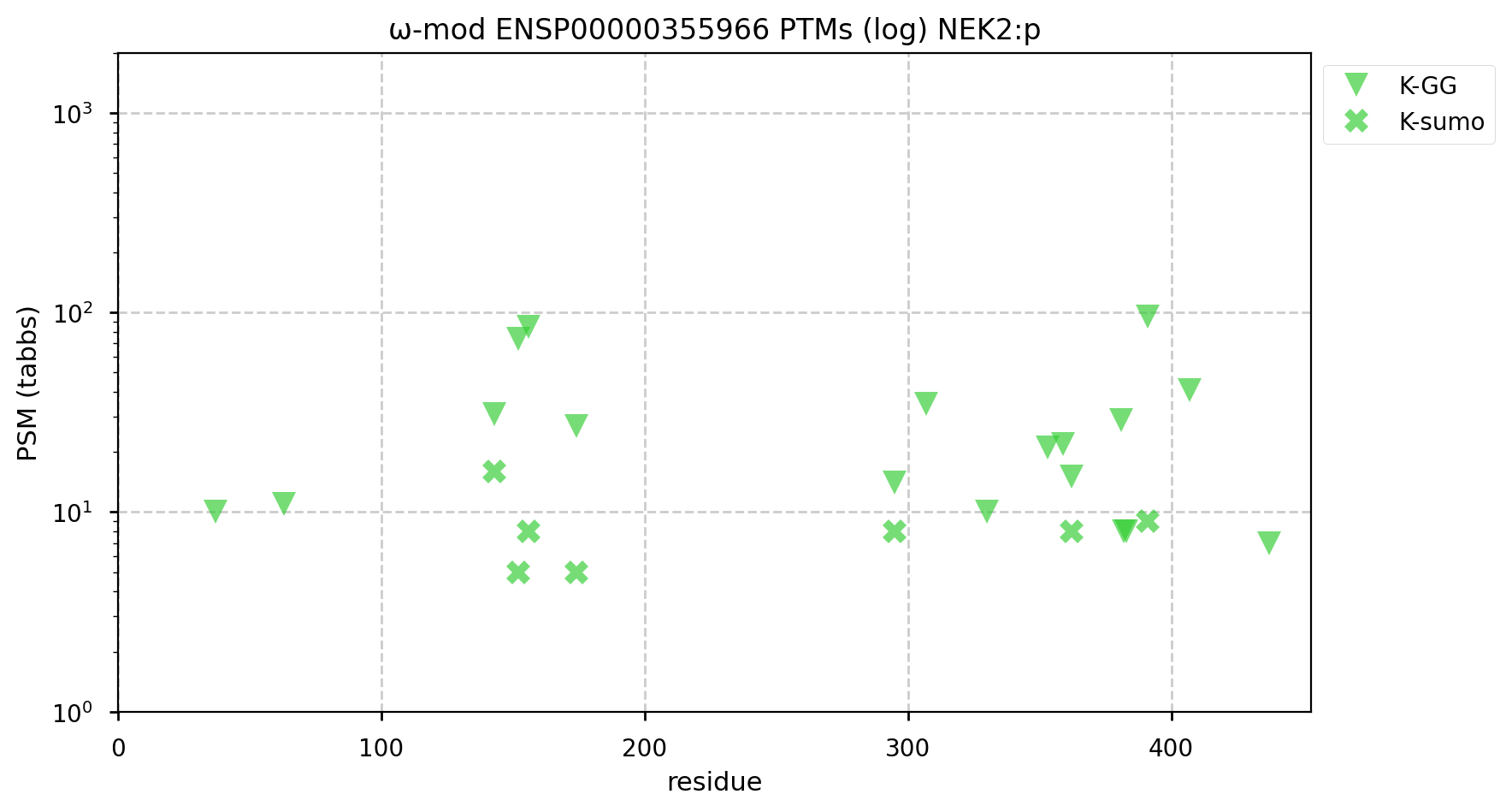
A GPMDB ubiquitination PTM diagram for human cyclin A2 (#CCNA2:p). Originally synthesized at the start of S phase, it is a Ub target of the anaphase-promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4944252). This is a less busy pattern of Ub acceptors than many proteins, but it seems to get the job done. #proteomics #anapc 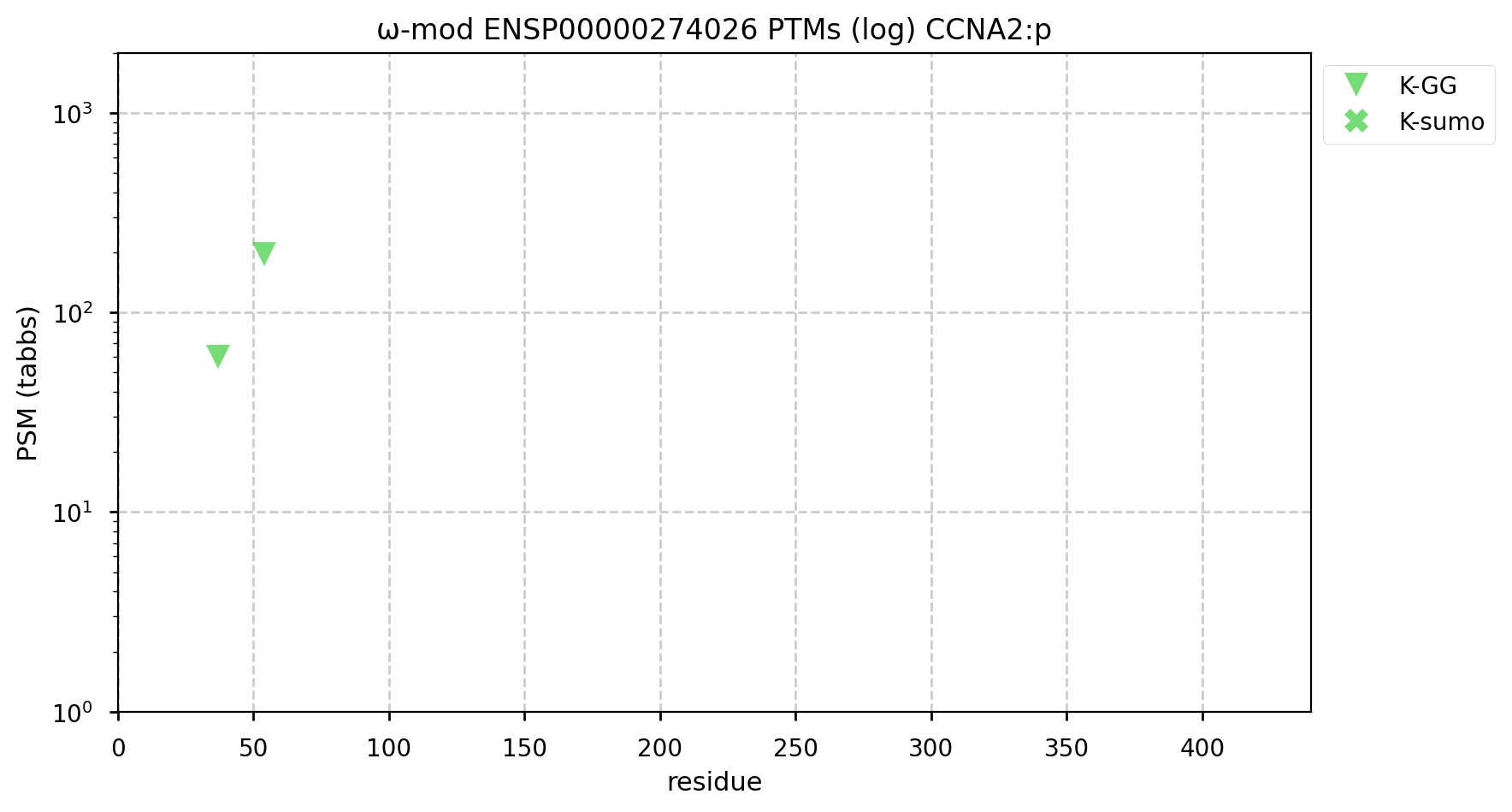
A GPMDB ubiquitination PTM diagram for human aurora kinase A (#AURKA:p). Involved in centrosome maturation & mitosis in general, it has been identified as a Ub target of the anaphase-promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6927175). #proteomics #anapc 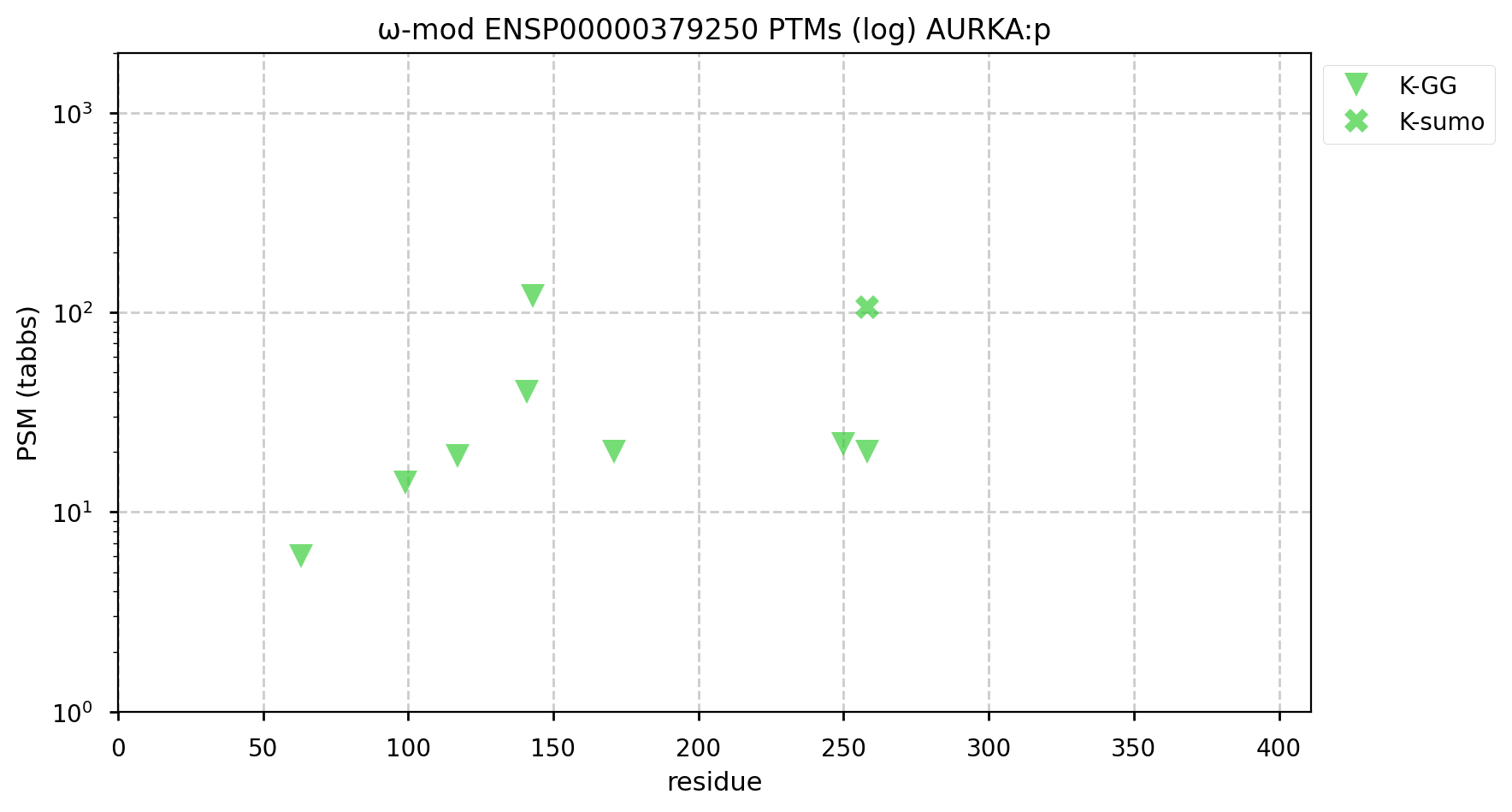
A GPMDB ubiquitination PTM diagram for human aurora kinase B (#AURKB:p). Involved in microtubule coordination during mitosis, it has been identified as a Ub target of the anaphase-promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7467358). #proteomics #anapc 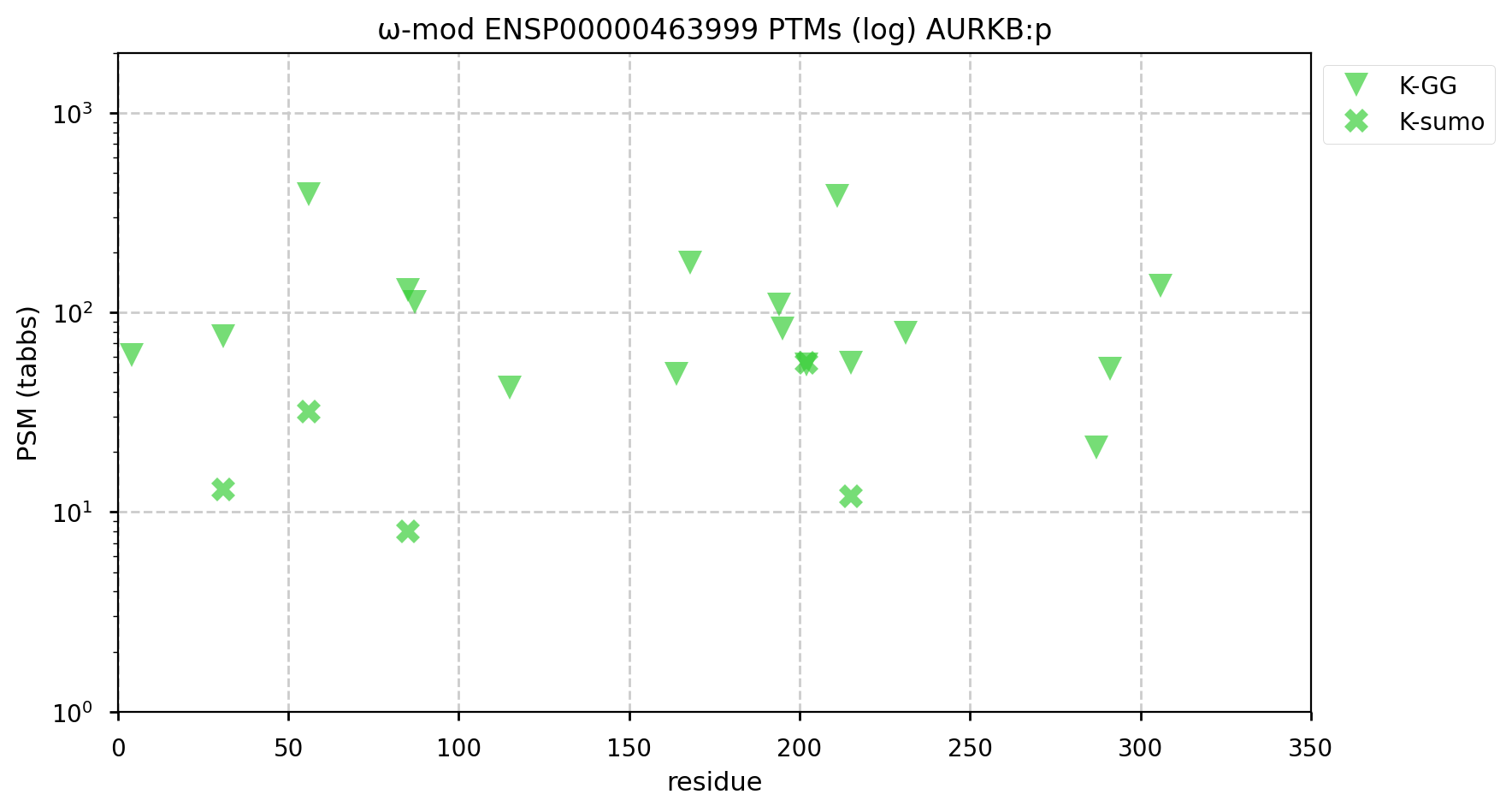
A GPMDB ubiquitination PTM diagram for human polo like kinase 1 (#PLK1:p). This kinase is produced during G1. It is part of the mitosis coordination system that has be ubiquitylated by the anaphase promoting complex and removed for the process to proceed to completion. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7467358). #proteomics #anapc 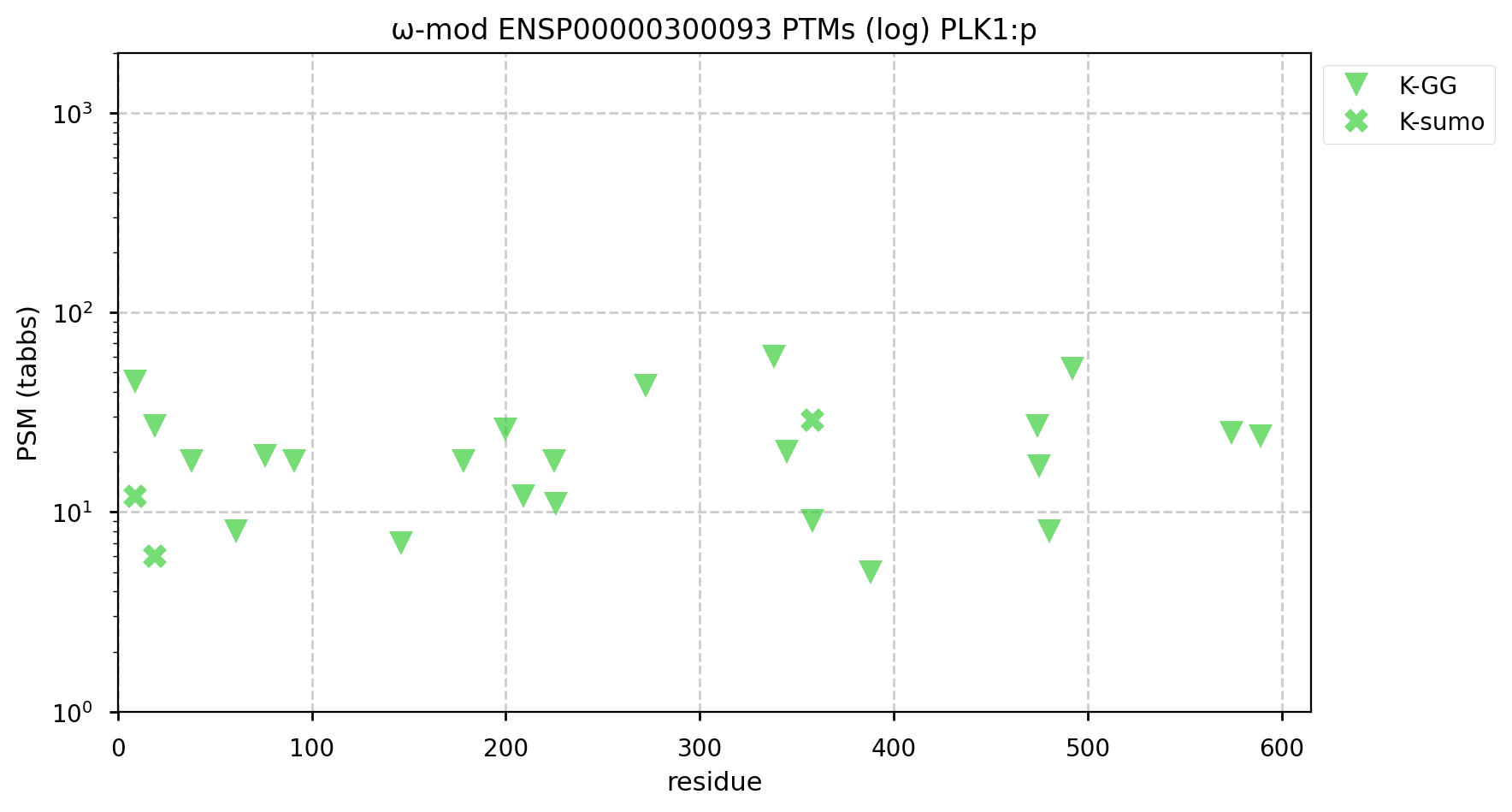
A GPMDB ubiquitination PTM diagram for human Targeting Protein for Xenopus plus end-directed kinesin-like protein 2 (#TPX2:p). There are ubiquitination acceptors on this sequence that the 'zines suggest are substrates for anaphase promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1291225/), it would appear that SUMOylation is doing some heavy lifting with this one, too. #proteomics #anapc 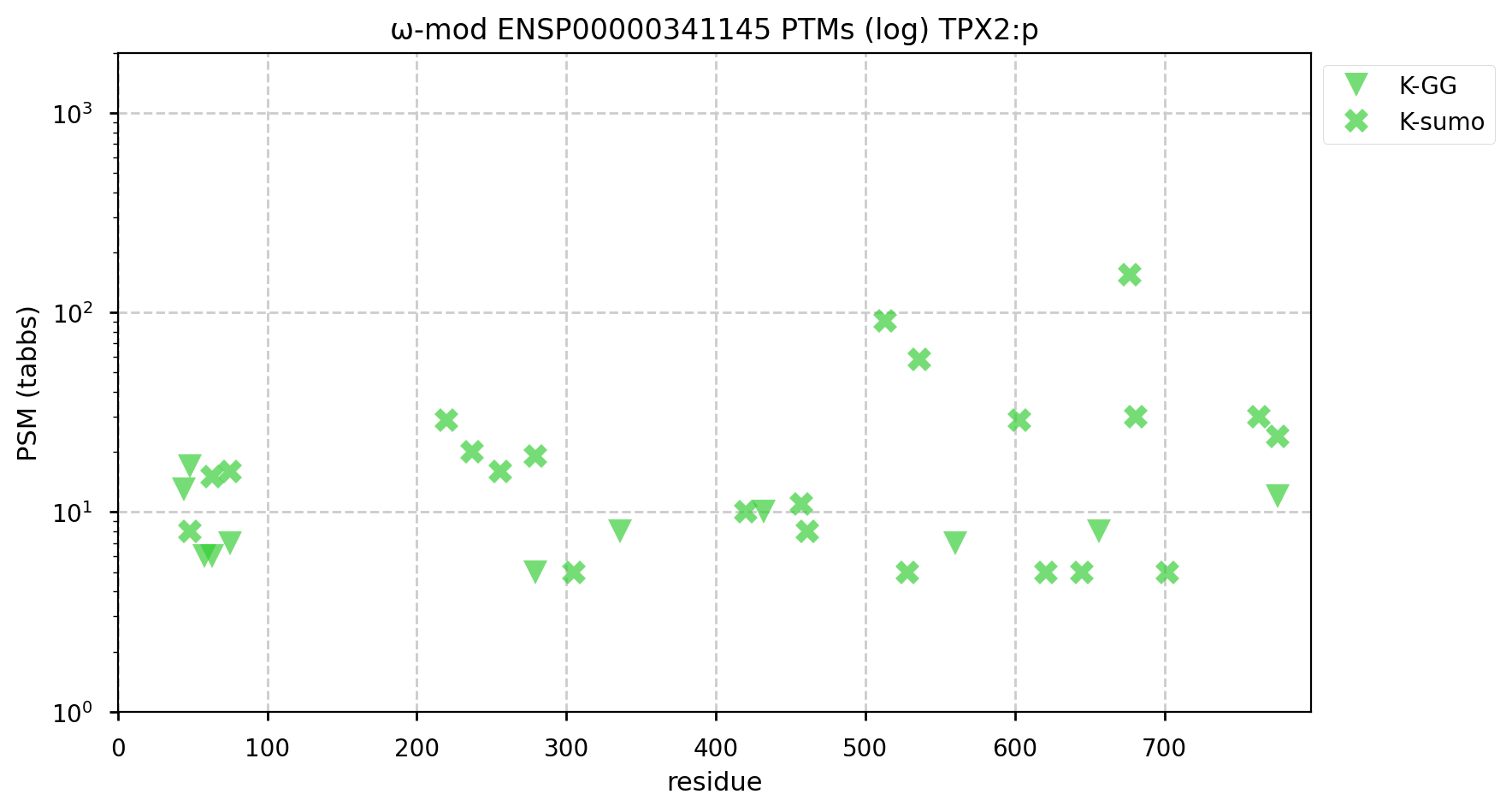
A GPMDB ubiquitination PTM diagram for human tumour protein p63 (#TP63:p). The 'zines are pretty adamant that the protein is a Ub substrate of the anaphase promoting complex (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6003458/). It has not been observed to be afflicted with SUMOylation. #proteomics #anapc 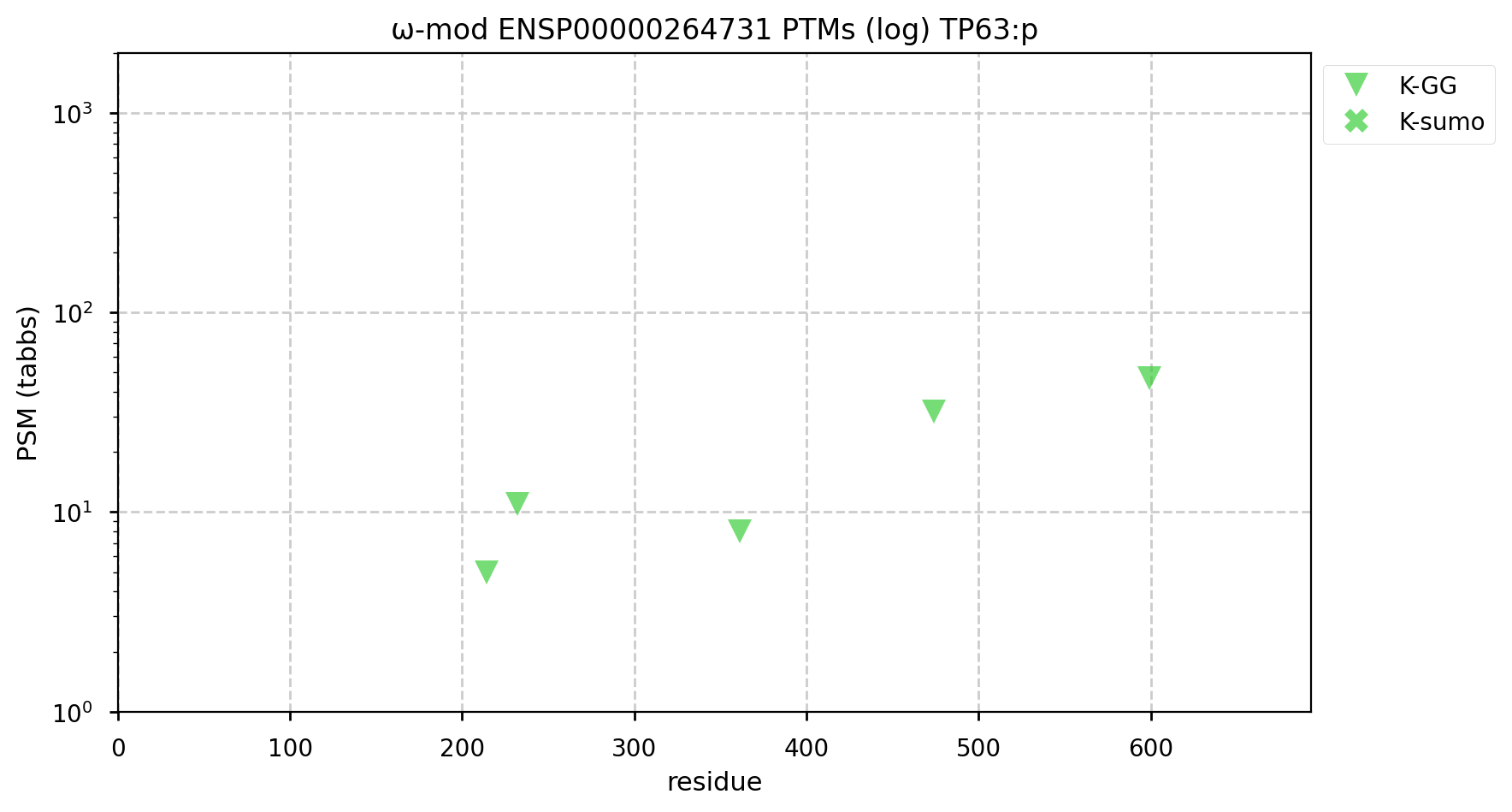
Further info:
After the origin recognition complex (ORC) has bound to the multiple origin
of replication sites on each chromosome, the minichromosome maintenance
(MCM) complex attaches to the ORCs. The presence ORC+MCM is necessary for
the initiation of DNA replication, i.e. the start of S phase,
inwhich the cell moves from G1's diploid chromosome count to G2's
tetraploidy. The following project compares the S/T phosphorylation patterns
of the six subunits of the heterohexameric MCM complex.
It is worth noting that the name "minichromosome maintenance" refers
to the genetic screen used in yeast that help identify the role of these
proteins in the DNA duplication process. Mammalian cells do not normally
contain minichromosomes & the MCM complex is a necessary component of the
cell cycle in eukaryotes.
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 2 (#MCM2:p). All three species have an N-terminal phospho-IDR with several high occupancy acceptors in at least 2 clusters. #proteomics #mcms 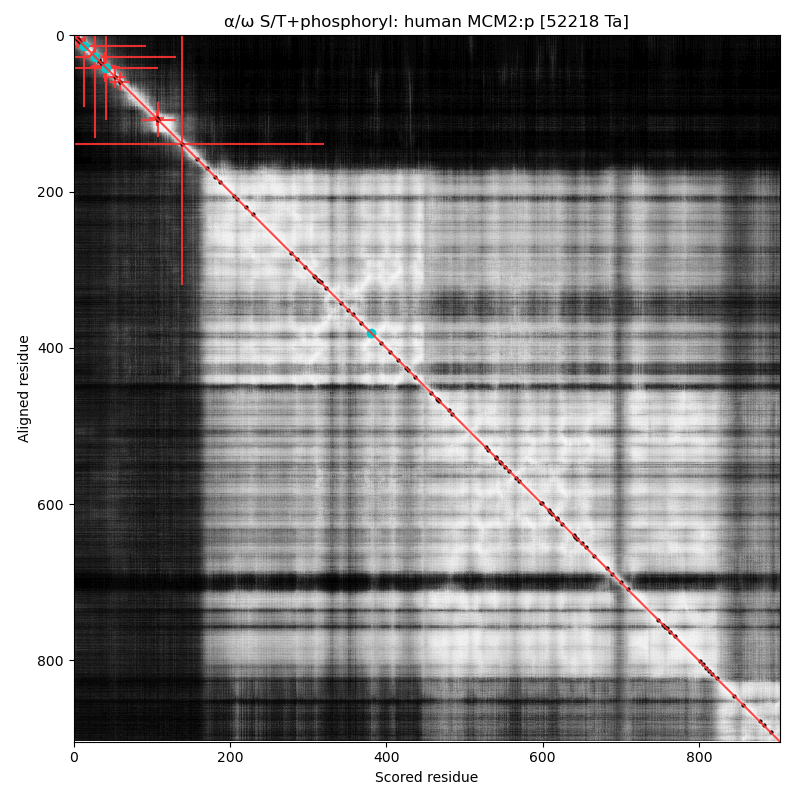 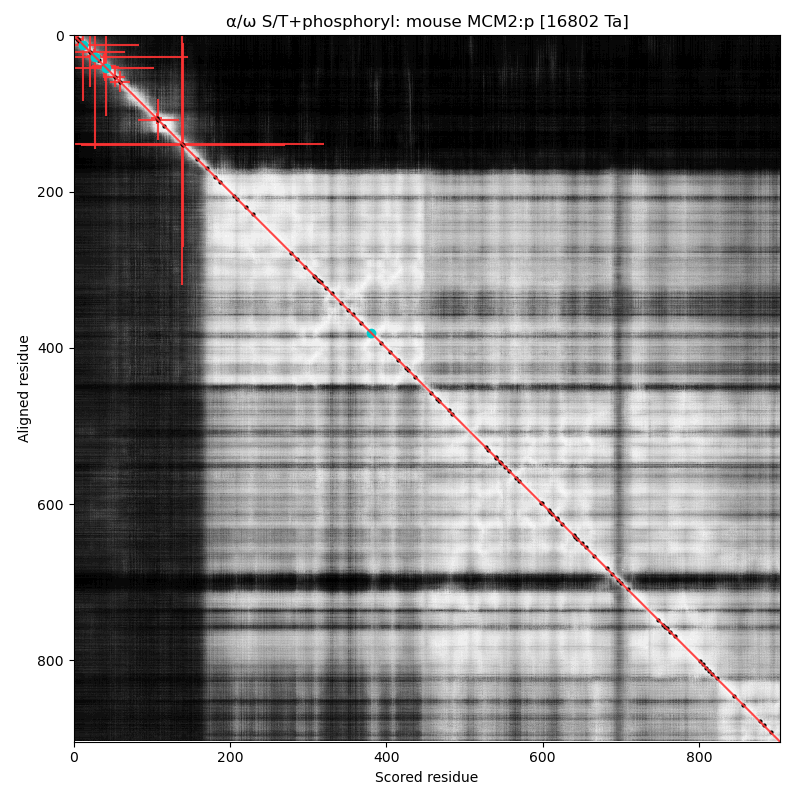 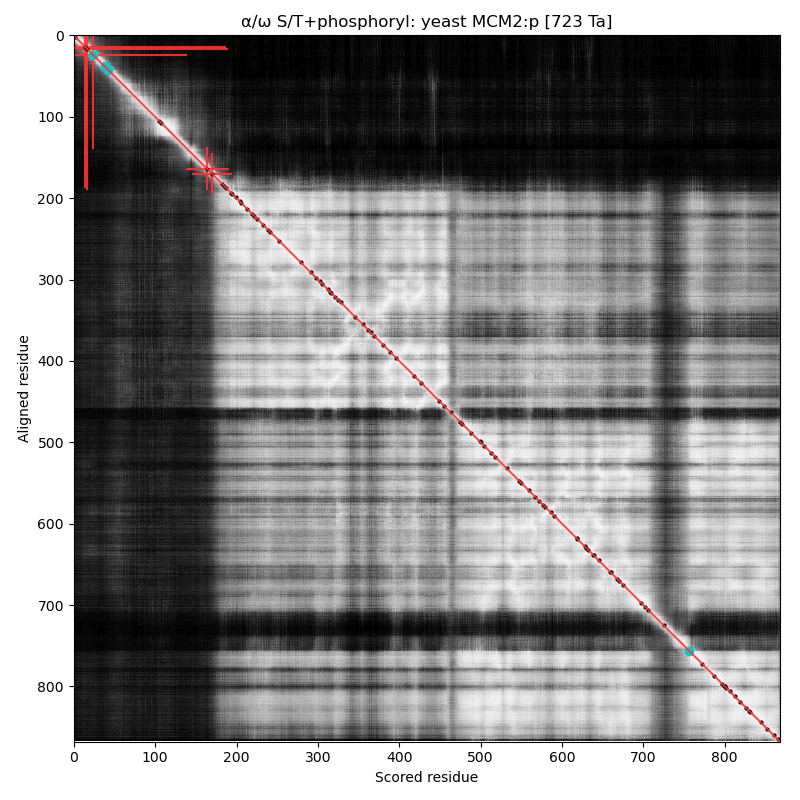
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 3 (#MCM3:p). All three species have a cward phospho-IDR with several high occupancy acceptors in at least 2 clusters. It is snuggled in between a composite of domains nward and a smaller unnamed C-terminal domain with a mix of helices and strands. #proteomics #mcms 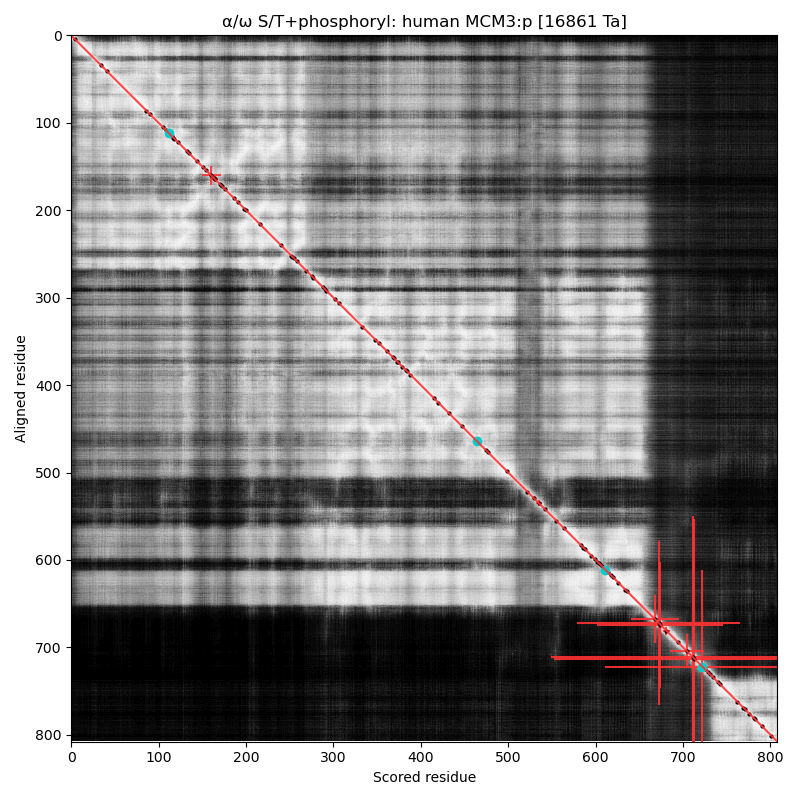 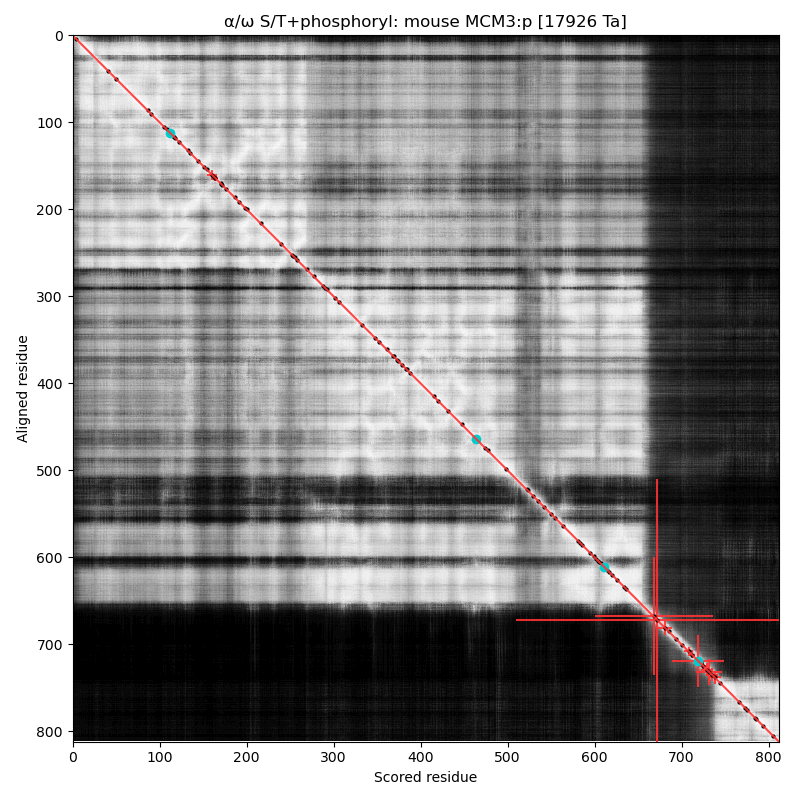 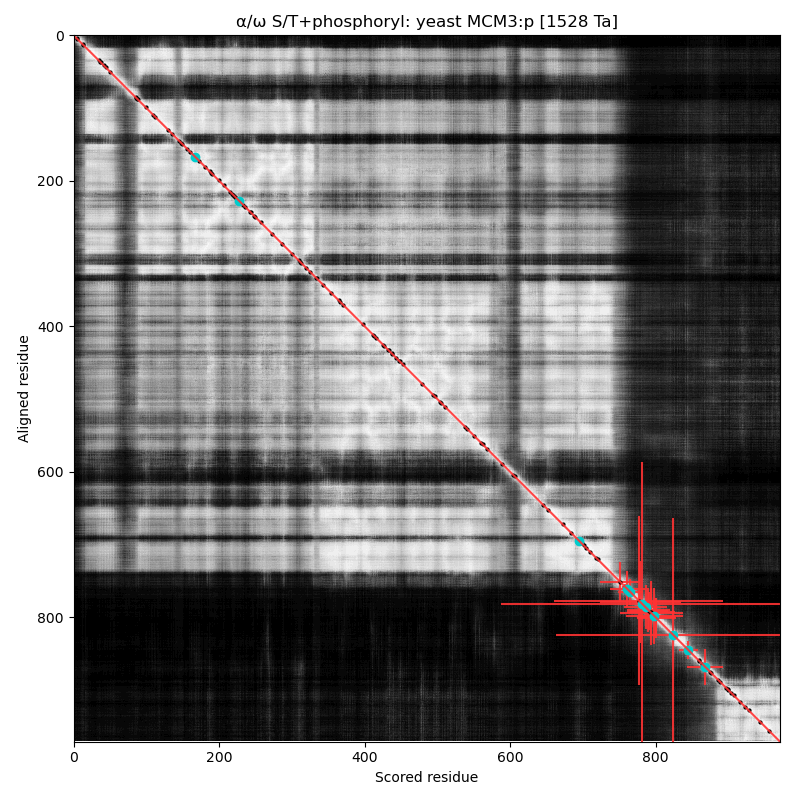
The AF diagrams for yeast and human MCM3:p, rotated to have the phospho-IDR noodle on the right, with the C-terminal shiny domain bottom right. 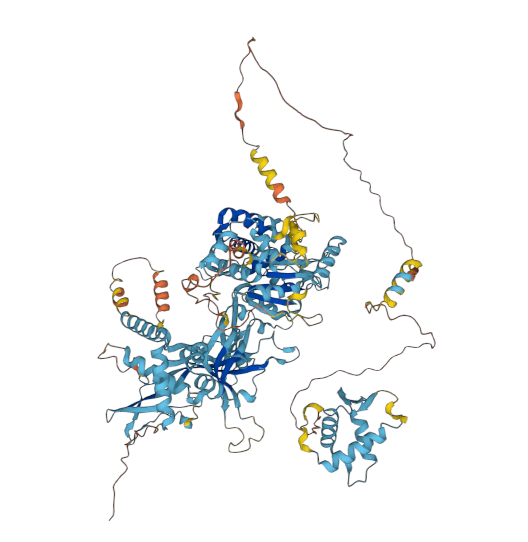 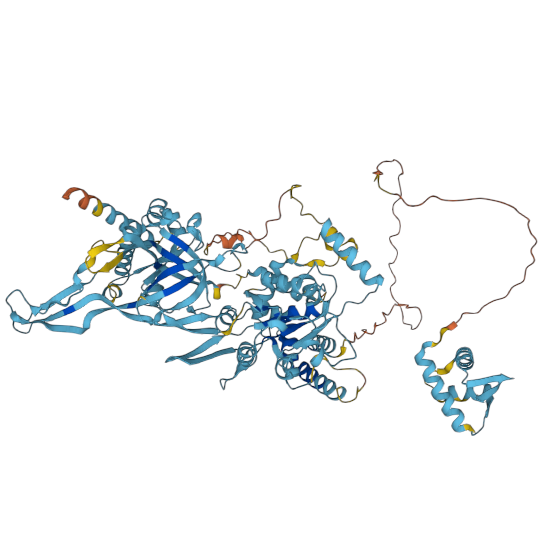
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 4 (#MCM4:p). All three species have a single phospho-IDR with high occupancy acceptors coming up like weeds between the N-terminus and the not-very-imaginatively named MCM domain. #proteomics #mcms 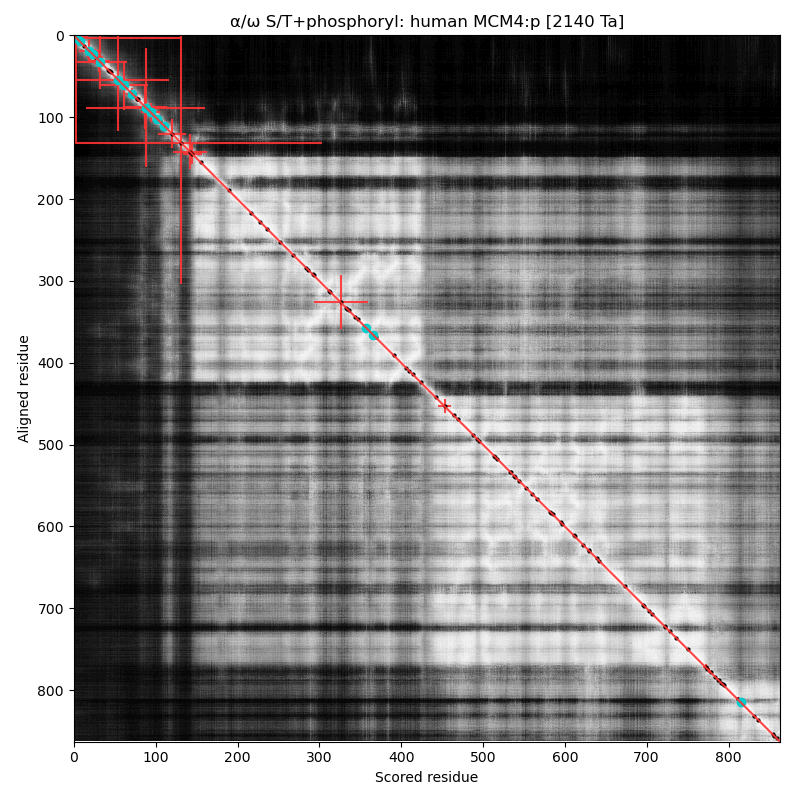 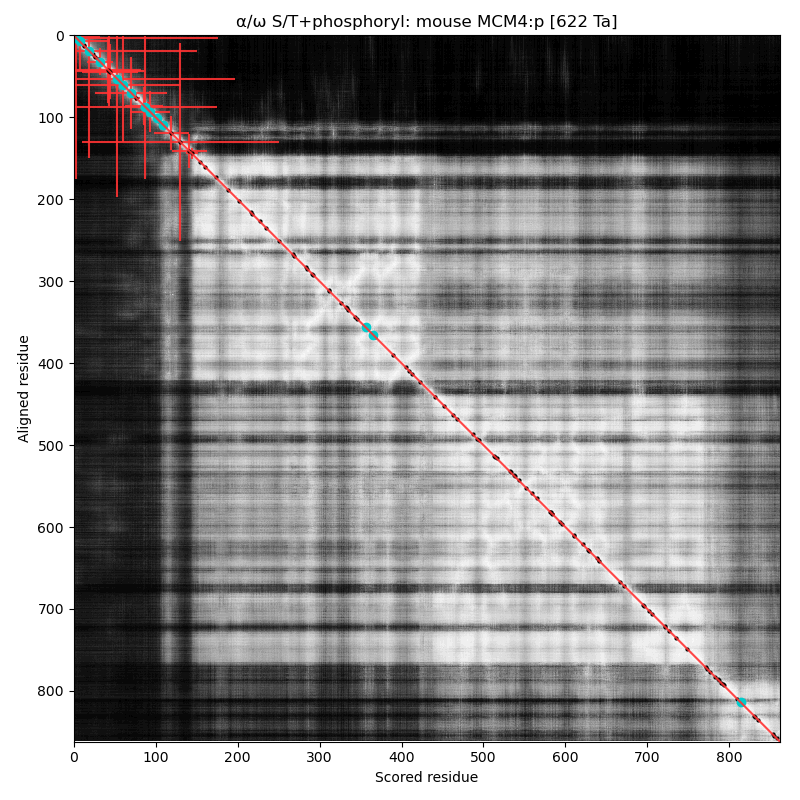 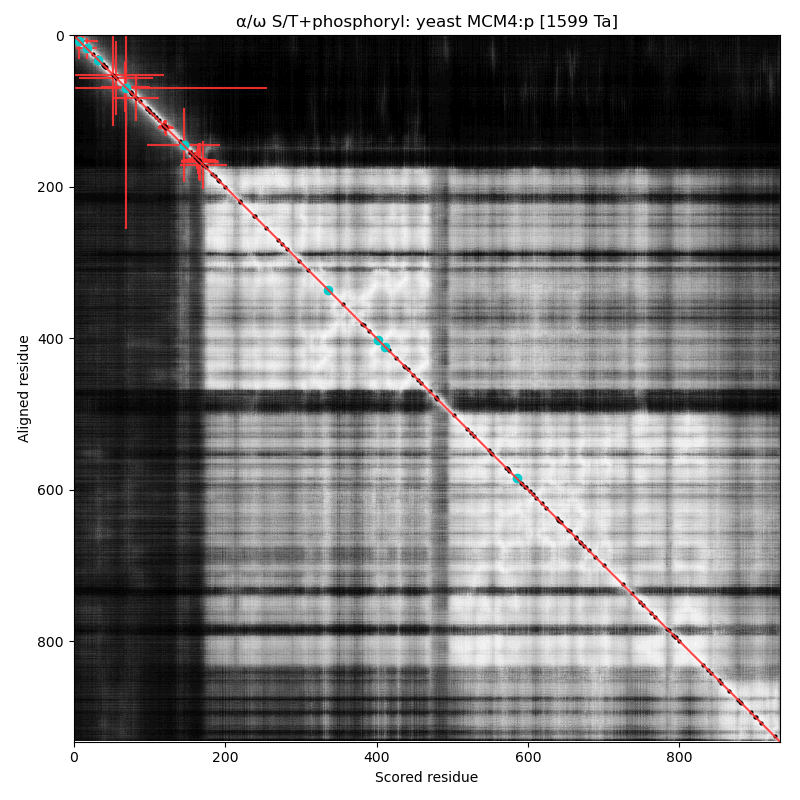
The yeast and human MCM4:p AF models, rotated so the phospho-IDR is on the left side of the image. 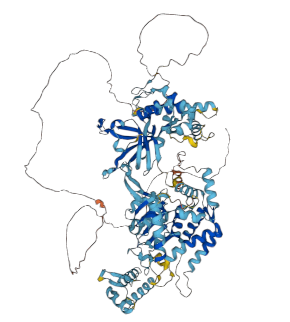 
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 5 (#MCM5:p). All three species have a single narrow N-terminal phospho-IDR each with 2 low occupancy acceptors clinging to the edge of the sequence. #proteomics #mcms 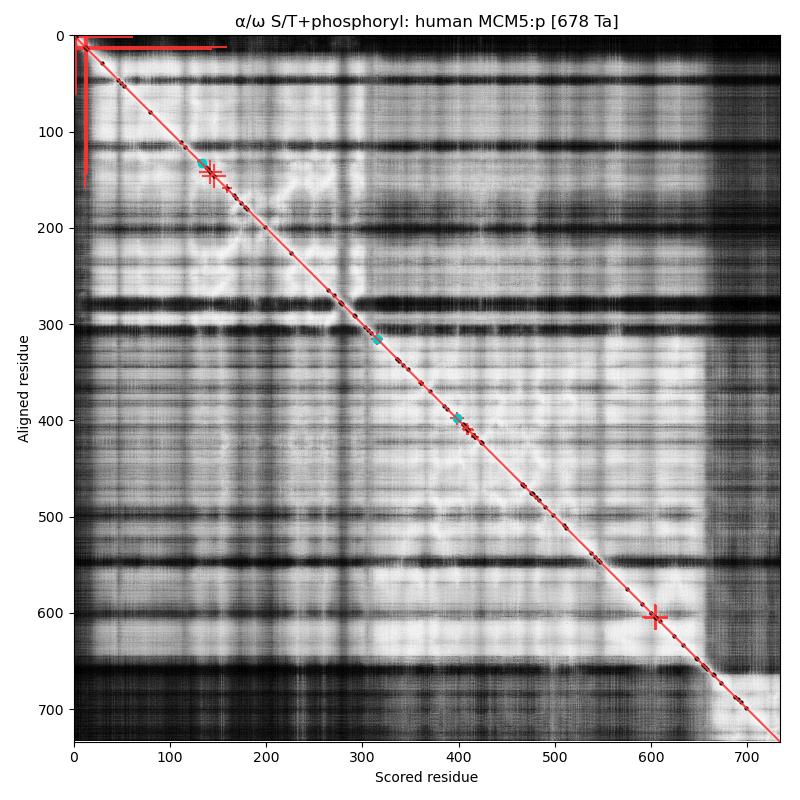 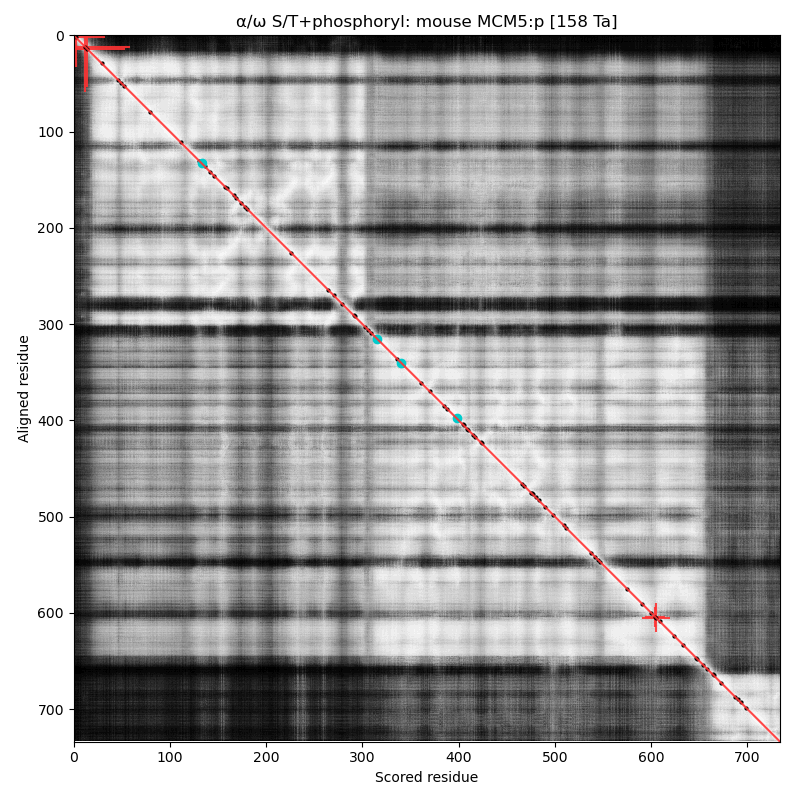 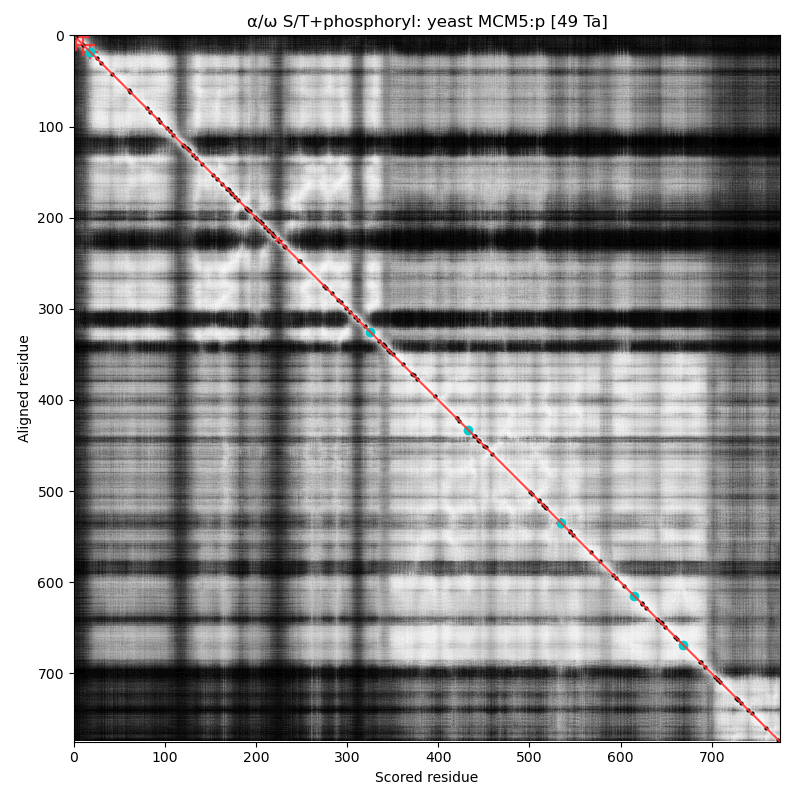
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 6 (#MCM6:p). The human & mouse patterns rhyme, although they have different emphasis on particular acceptor clusters. Yeast stands alone. #proteomics #mcms 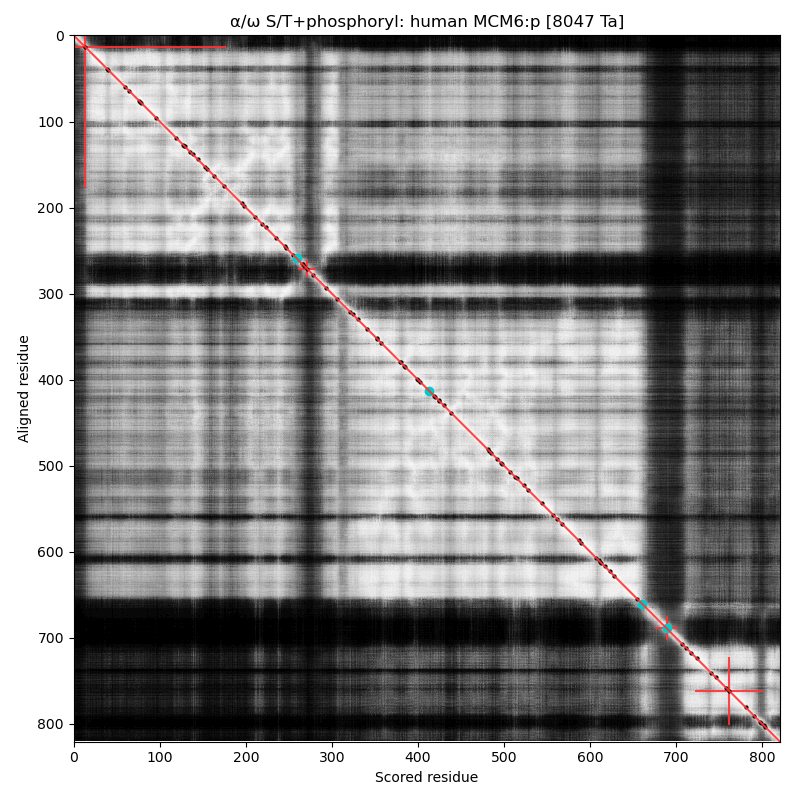 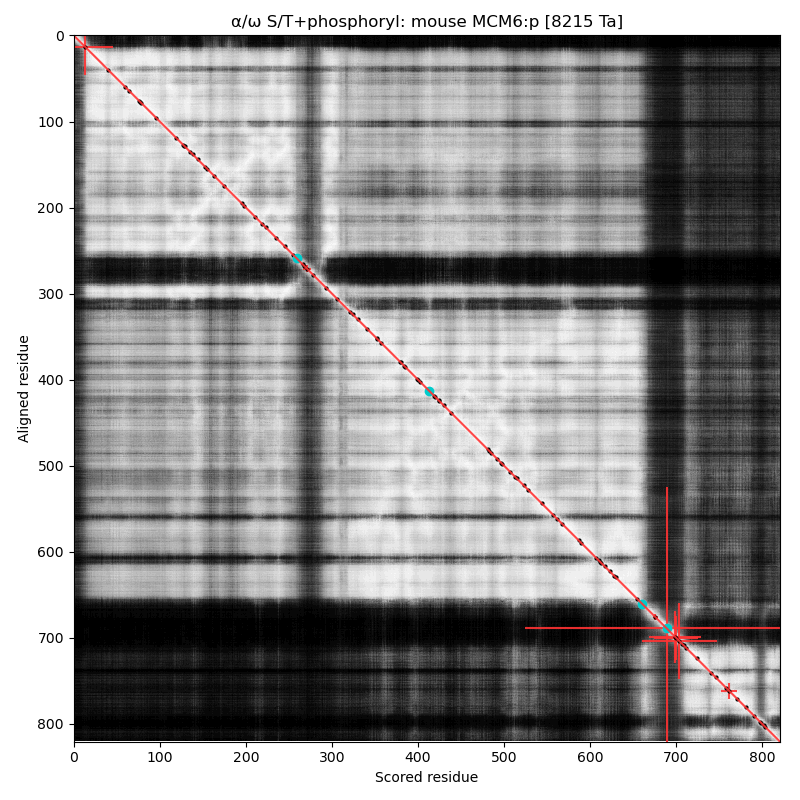 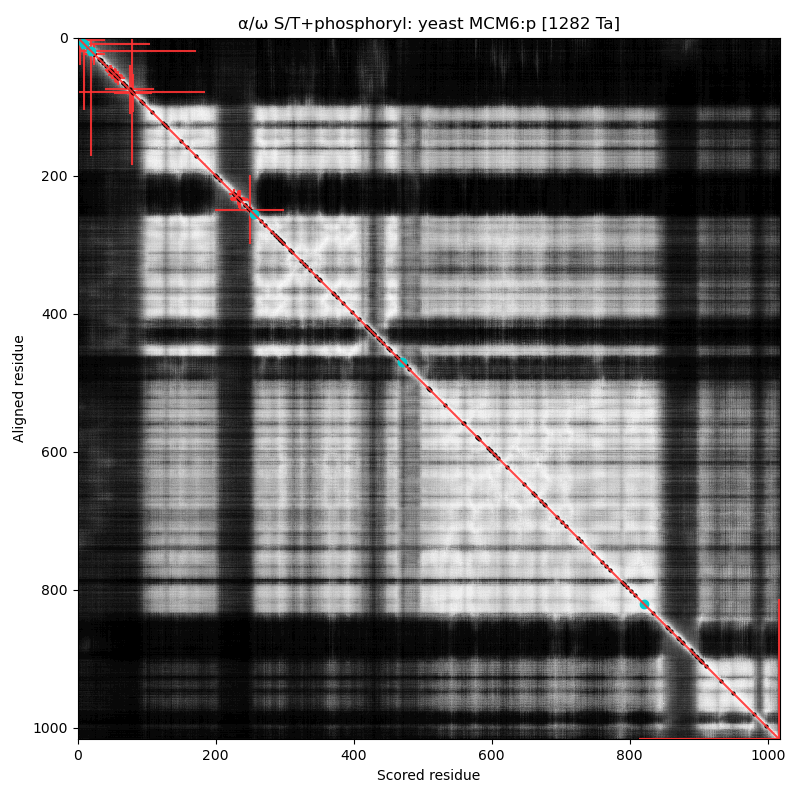
α⧸ω ST phosphorylation diagrams for human, mouse & yeast minichromosome maintenance complex subunit 7 (#MCM7:p). The human & mouse patterns have low occupancy acceptors in narrow IDRs connecting helices. As with MCM6:p, yeast stands alone. #proteomics #mcms 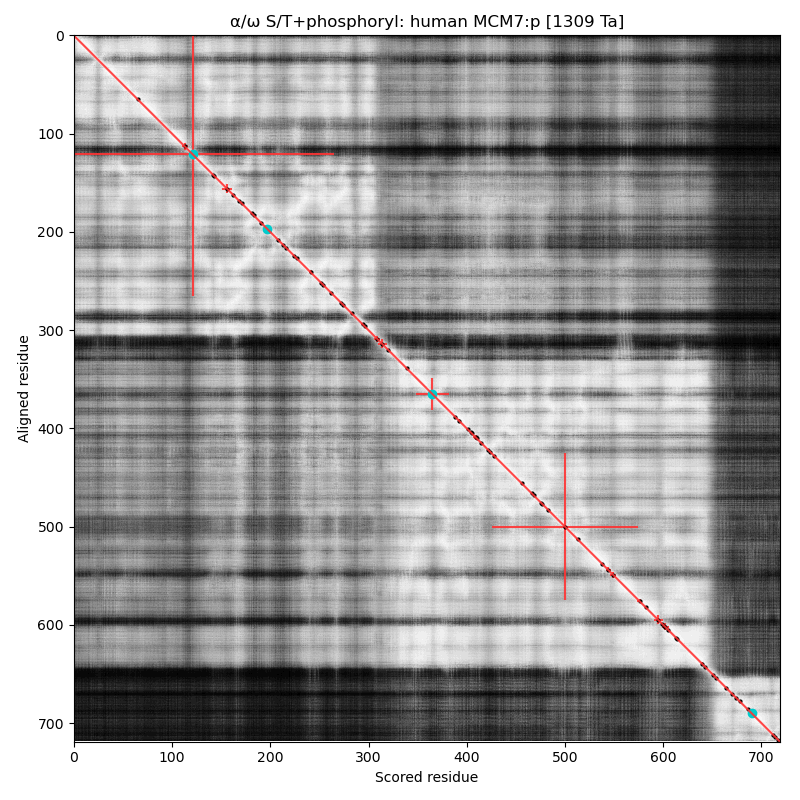 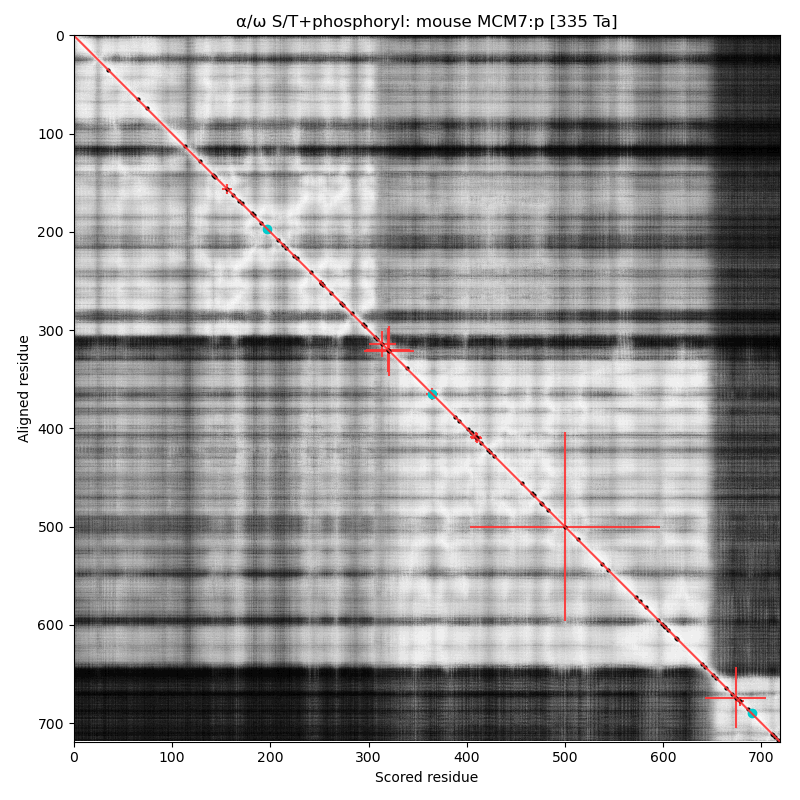 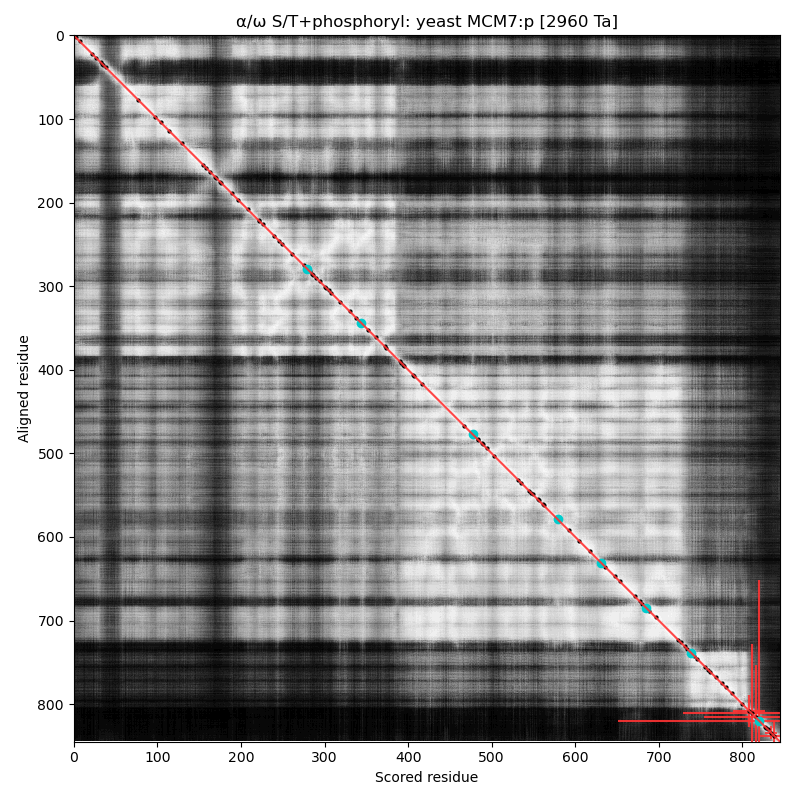
The origin recognition complex (ORC) is necessary to initiate the duplication of DNA during cell cycle:
if it is defective, cells stall in G1. Discovered in yeast, all cells use a very similar
complex in their cell cycles. This little project shows diagrams of the occupation of S/T phosphorylation
acceptors in the complex's subunits in the human, mouse & yeast proteomes, with one posted daily
over 6 days.
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 1 (#ORC1:p). The origin recognition complex is a 6 subunit assembly that becomes active in the transition from G₁ to S phase & is necessary for the initiation of DNA replication. Phosphorylation of this particular subunit is on the IDR between the nward bromo adjacent homology & the cward AAA+ ATPase domain in all 3 species, but the relative positions of the occupied acceptors vary. #proteomics #orcs 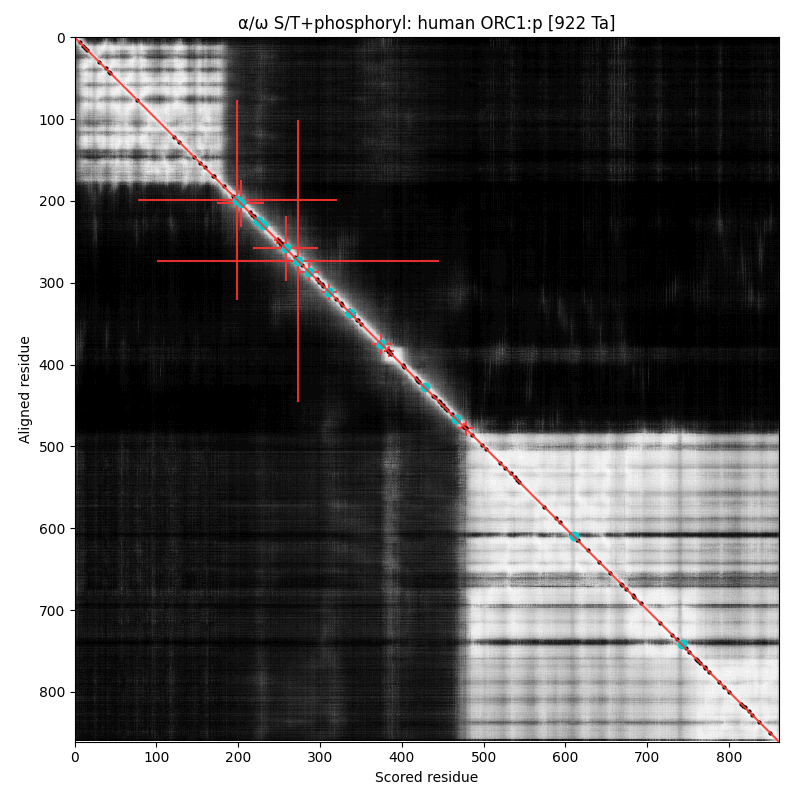 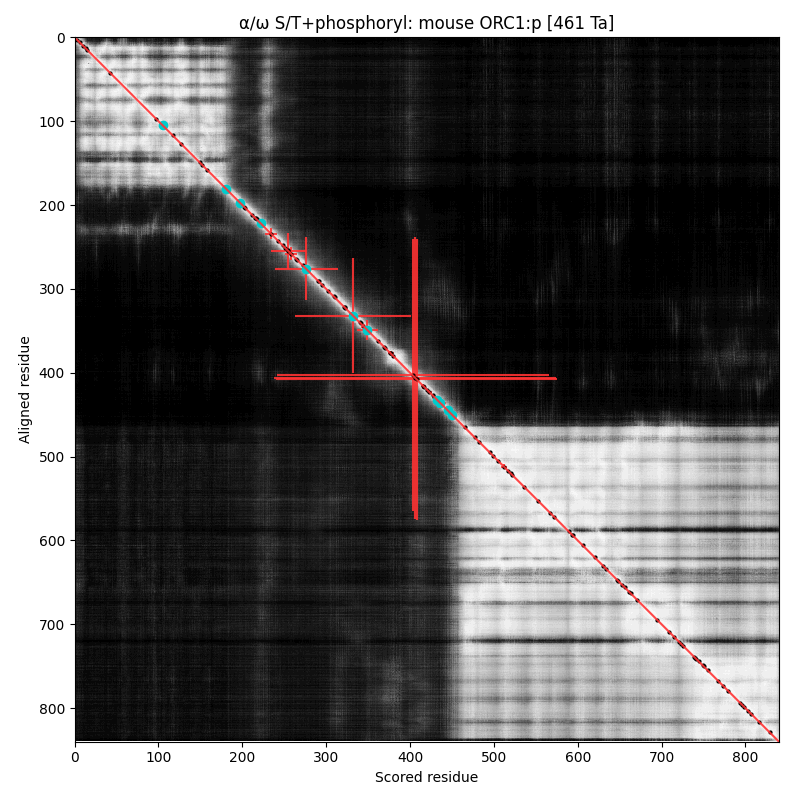 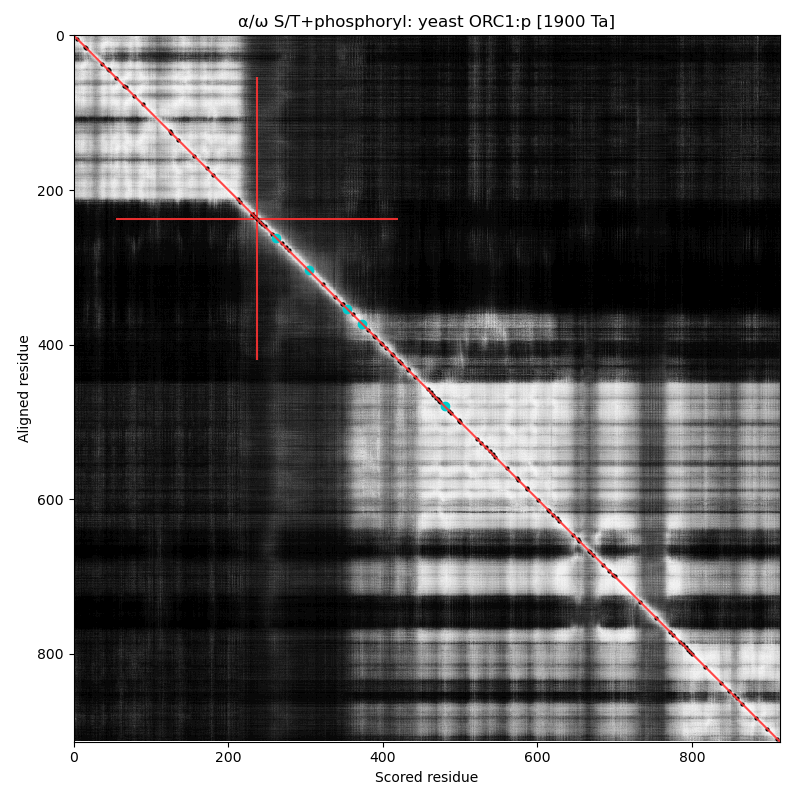
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 2 (ORC2:p). In 2 out of 3 species there are some occupied acceptors in the nward IDR, but mouse seems to do OK with next to nothing observable in this region. #proteomics #orcs 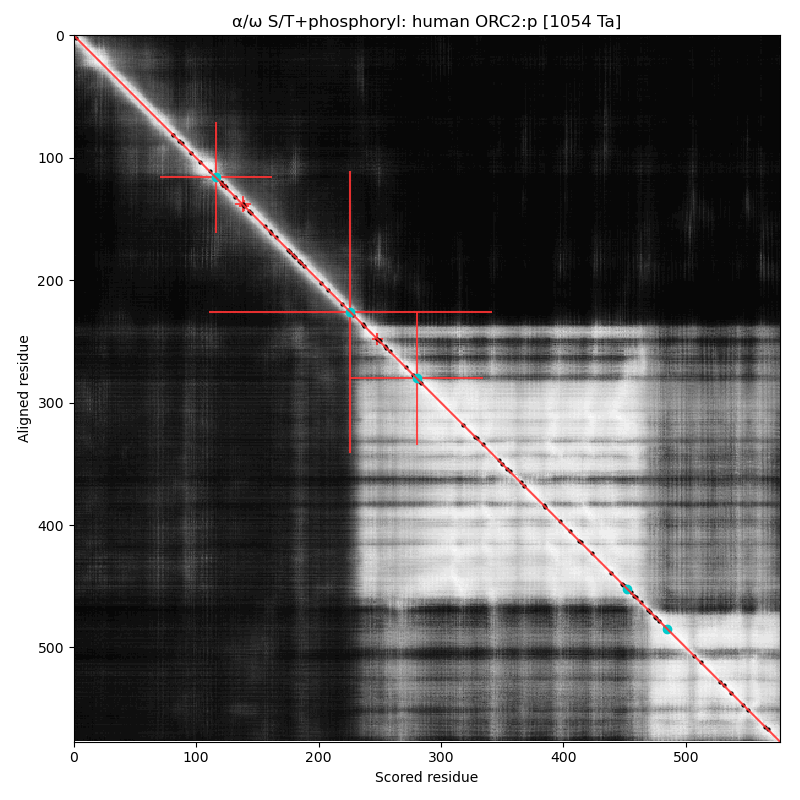 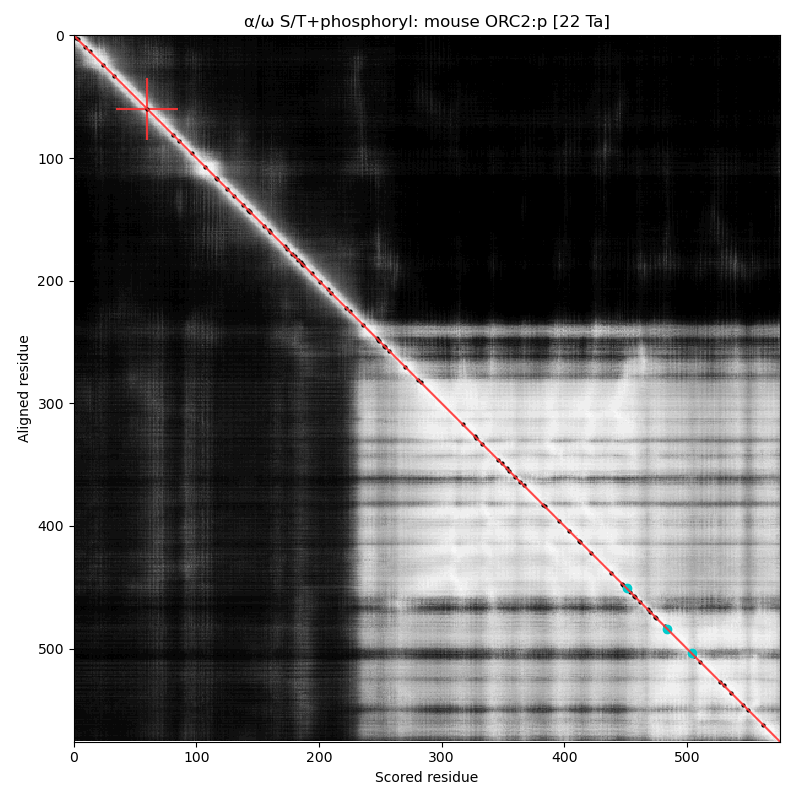 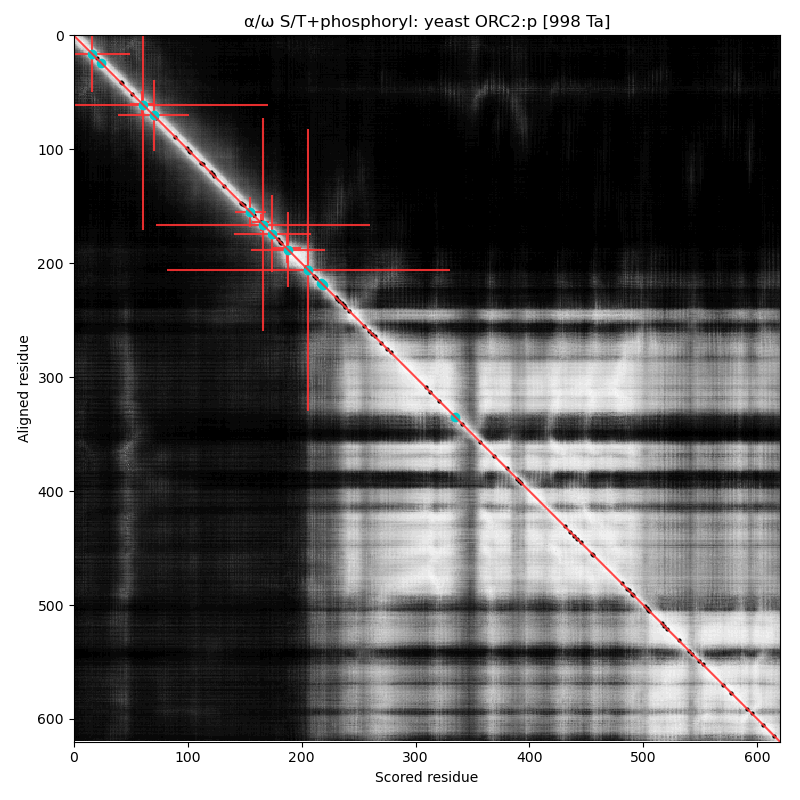
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 3 (#ORC3:p). The human sequence has several low occupancy acceptors in narrow IDRs, while both mouse and yeast have no observed phosphorylation. #proteomics #orcs 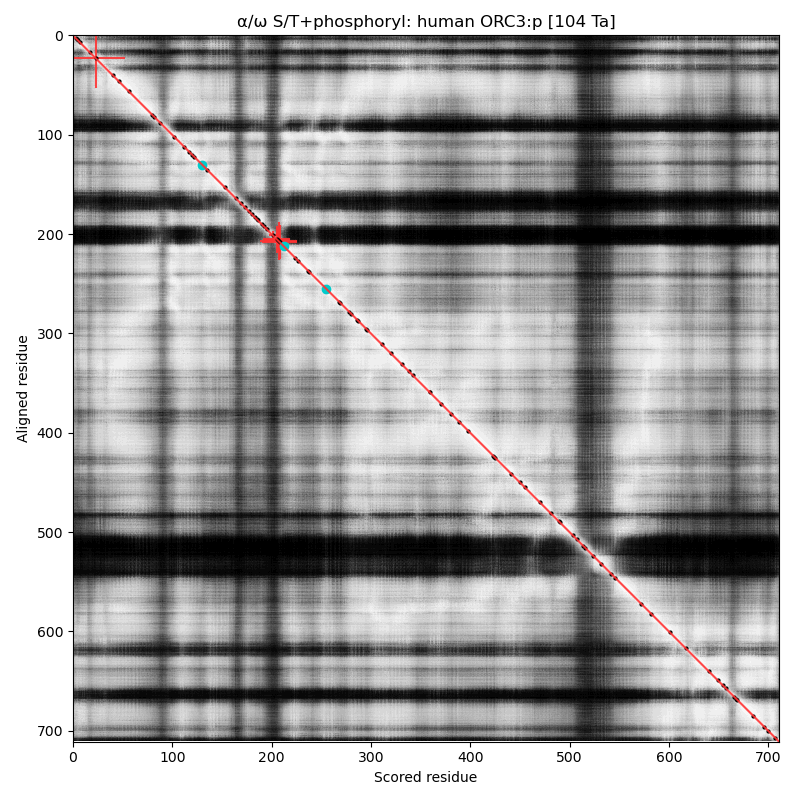 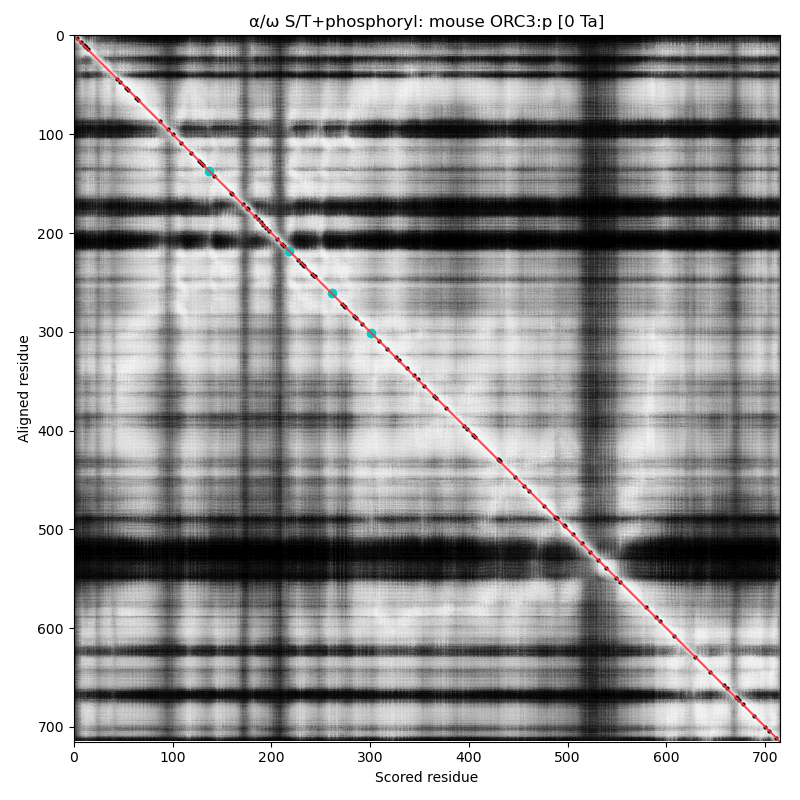 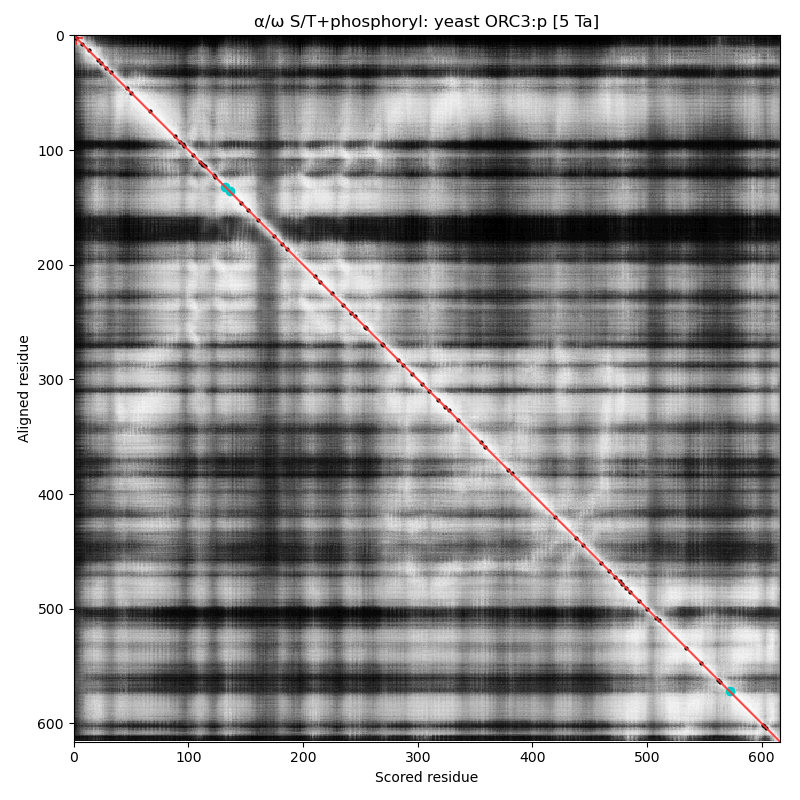
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 4 (#ORC4:p). The all 3 sequences have a single, low occupancy acceptor in a narrow IDR at the N-terminus. #proteomics #orcs 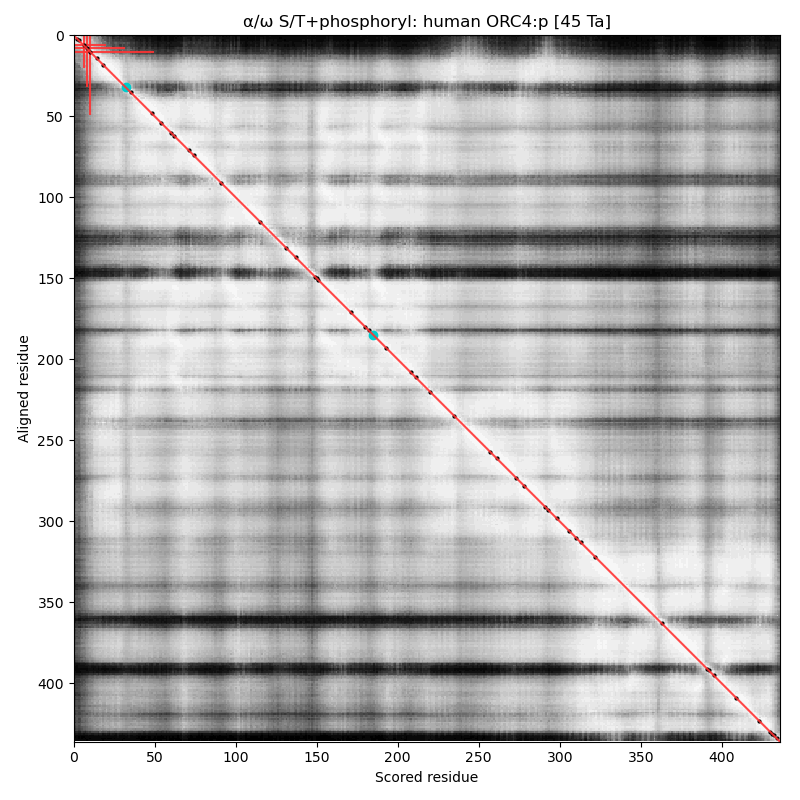 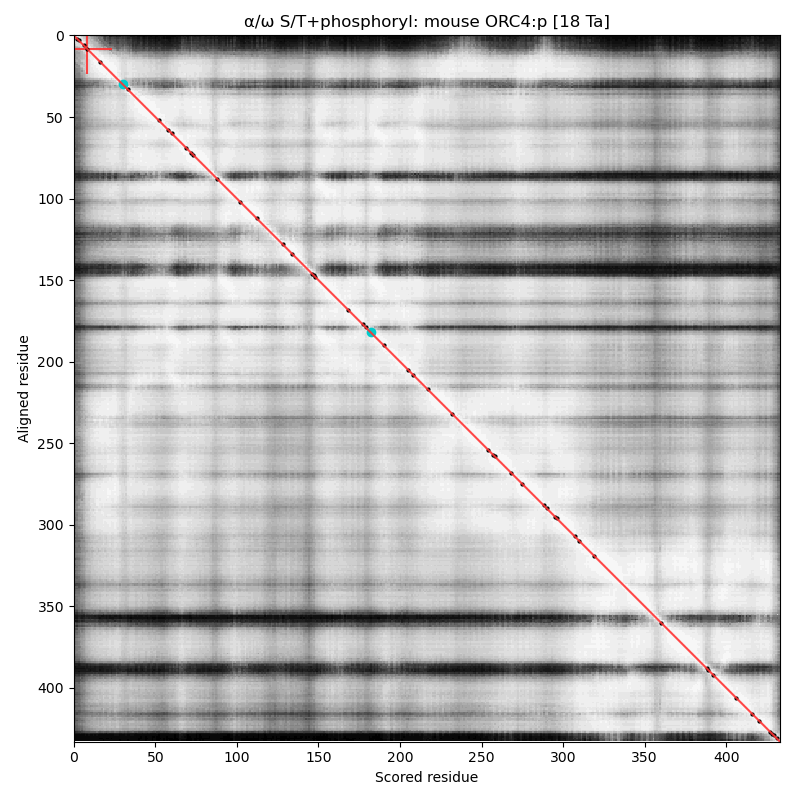 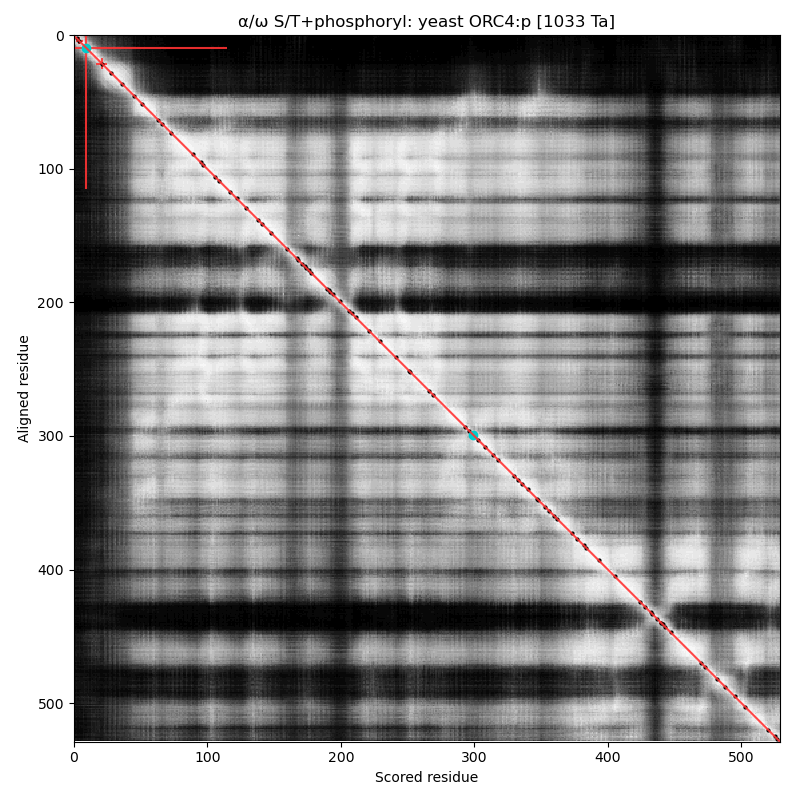
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 5 (#ORC5:p). This subunit doesn't have any significant phosphorylation: probably not coincidentally it also lacks any S/TP sites in its sequence. #proteomics #orcs 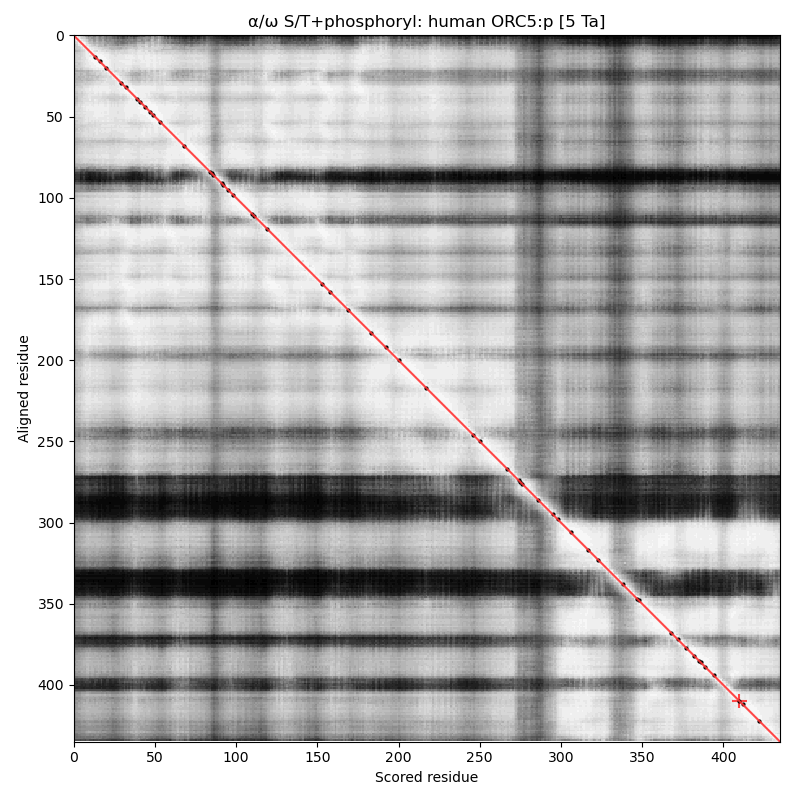 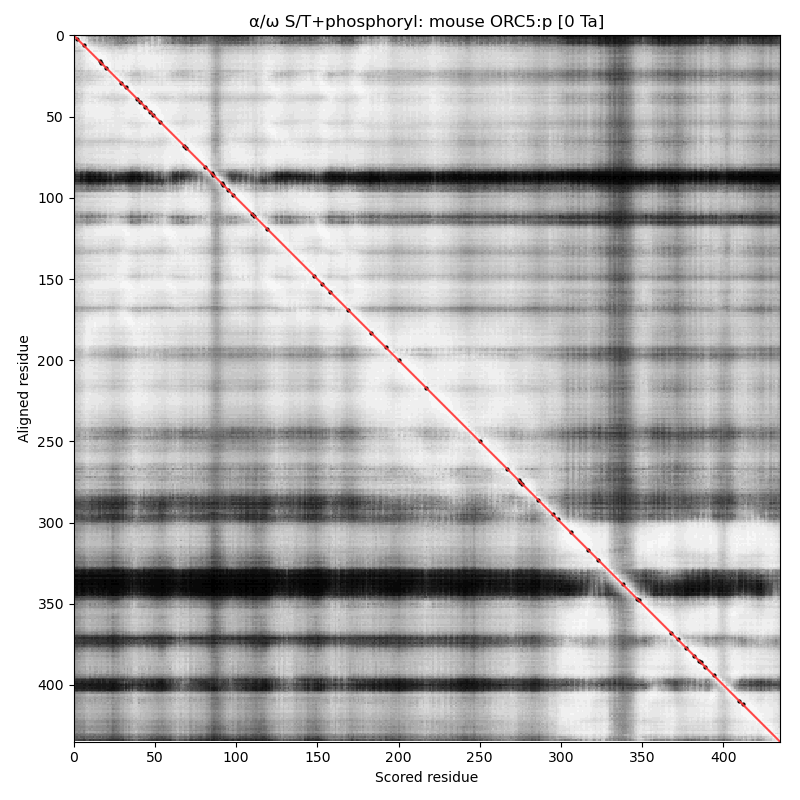 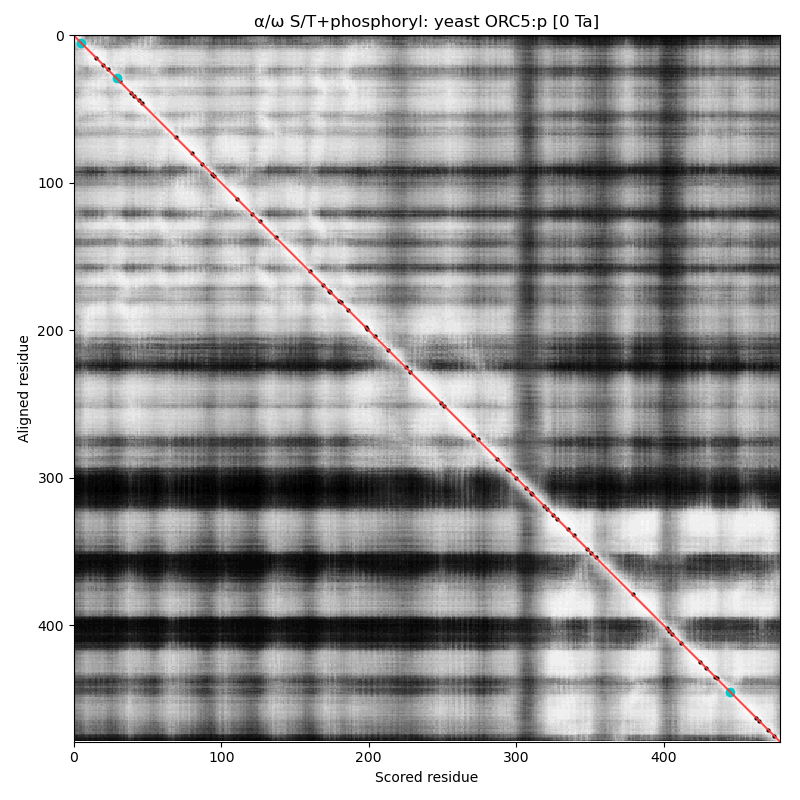
α⧸ω ST phosphorylation diagrams for human, mouse & yeast origin recognition complex subunit 6 (#ORC6:p). The mammalian subunits are very different than the fungal subunit. #proteomics #orcs 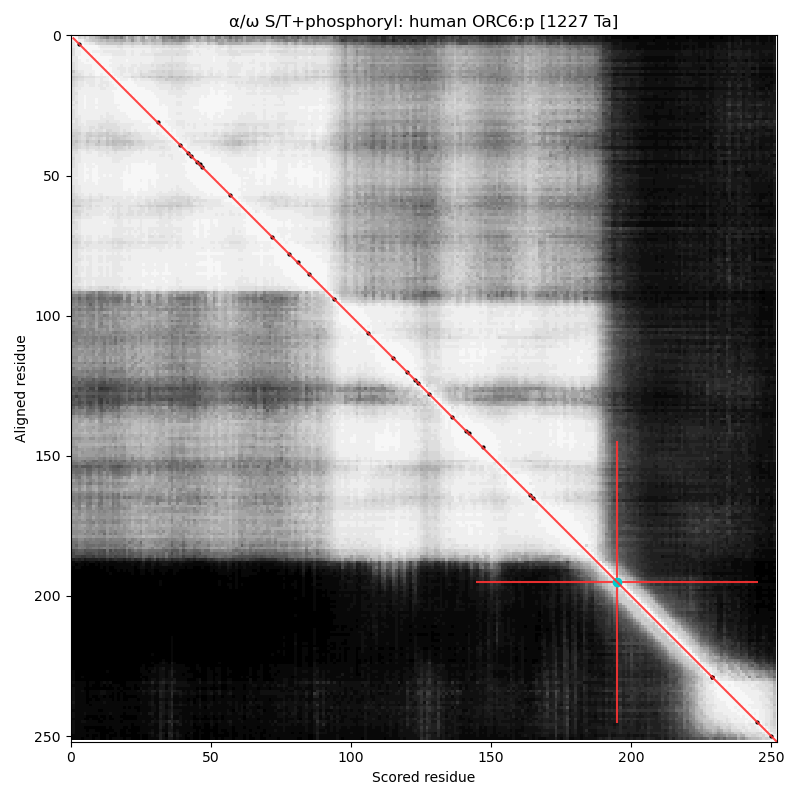 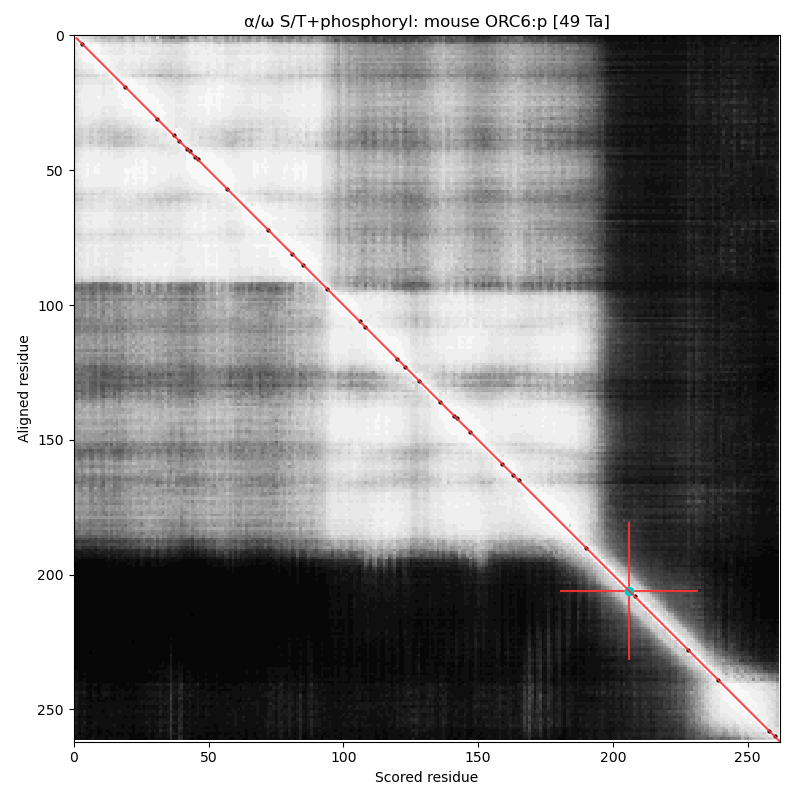 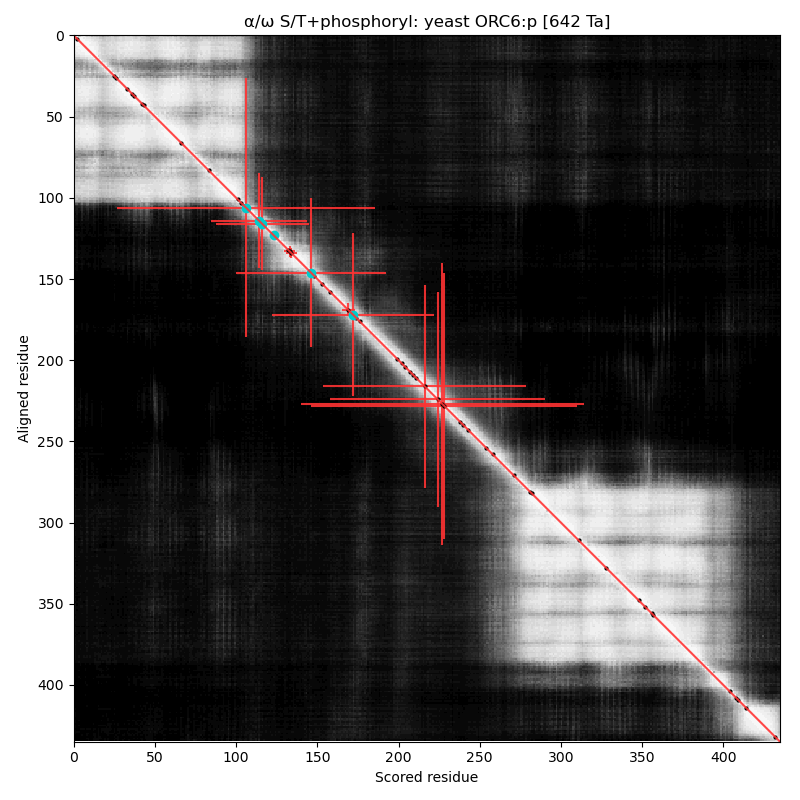
Copyright © 2024, The Global Proteome Machine.
Located at 137 Bannatyne | Privacy Statement
|