|
|
|
Data set of the week: (2009/12/28)
Community proteogenomics reveals insights into the physiology of phyllosphere bacteria
This dataset was transfered to GPMDB via ProteoExchange from PRIDE (see data).
It is credited to Delmotte N, et al. and it is described in Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16428-33.
Data-set-of-the-week is a new feature for GPMDB, started with the intent of highlighting
high quality data sets that have been made available via GPMDB and ProteomExchange. Data
sets will be selected by a panel, but any suggestions (email to dsotw@thegpm.org) of suitable data will be
considered.
|
|
The 1,000 most observed human proteins
(2009/11/06)
This spreadsheet (human_top_1000.xls)
is a list of protein sequences that have been observed most often by GPM users who used the "human"
GPM search server. The columns in the spreadsheet are as follows:
- Column A: ENSEMBL protein accession number for the sequences;
- Column B: HUGO Gene Naming Committee symbol for the associated gene;
- Column C: NCBI gene number for the associated gene;
- Column D: International Protein Index accession number for the sequence;
- Column E: SwissProt/Uniprot accession for the sequence;
- Column F: the probability that a protein will be found in a dataset (as a percentage);
- Column G: the base-10 log of the minimum expectation value found for that protein; and
- Column H: a text description of the protein.
The value in Column F was calculated by taking the number of times (ni) that the protein was observed
in the approximately 24,000 (N) datasets examined and doing the simple calculation:
pi = 100(ni/N)
A "dataset" corresponds to a submitted set of MS/MS spectra, which results in a GPM result file, so it
is roughly equivalent to the set of data from an LC/MS/MS run. A protein can only be observed once in a dataset.
|
|
110,000,000th Peptide Id Recorded
(2009/11/02)
Over the weekend, GPMDB passed the 110 million mark for peptide
identifications. We would like to thank all of the data
contributors who have made this project a success. Special
thanks goes to our ProteomExchange partners TRANCHE and PRIDE,
for making their data available.
|
|
Service interuptions for maintenance (2009/08/14)
We are performing some long delayed maintenance on the computers in the GPM system. There may be some service
interuptions throughout the system starting Friday, August 14, lasting until Monday, August 17.
|
|
New data views: Protein-Protein Interactions and Groups (2009/07/20)
Two new views of GPM data sets have been added. The "ppi" view (available for ENSEMBL human and yeast accesssion numbers)
is similar to the existing pathways display. The new display categorizes all of the proteins found in a dataset with the
proteins in corresponding protein-protein interaction sets listed in BioGrid (human and yeast) and HPRD (human only).
The "group" view is simply a list of all proteins found as groups, with the primary member of a group
being the protein displayed in the main model view, along with all of the proteins that
could be obtained by using the individual homology lists. Any protein that has at least one spectrum assigned to a peptide sequence that is unique
for that protein is listed as a primary protein.
Additionally, a link has been added that generates an MGF formatted annotated peptide spectrum library from the
results of a single data set. This MGF file is formatted so that it can be used as a library for standalone instances of X! Hunter.
|
|
Please pickup your data (2009/05/12)
Last week, a user was very diligently attempting to get search results for
two data sets, from the data files RAT592_C-all_20090430_LT.mgf and RAT592_B-all_20090430_LT.mgf. Unfortuately, these
data files required more memory than was available on the search servers, so they failed to execute. We have
re-run the data and the results are now available as GPM77700007010 and
GPM77700007011 respectively. These data files
both contain over 200,000 original spectra and appear to be composed of spectra merged together from LC/MS runs on individual gel band slices, using
an ion trap instrument, from a sample that appears to be rat inner ear tissue.
|
|
New release of X! Hunter ASLs (2009/05/04)
The May 1st, 2009 release of the Annotated Spectrum Libraries — used by X! Hunter for high speed, high accuracy protein
identification — is now available here. This new release contains libraries for commonly used eukaryote species as well as three SILAC libraries
and libraries for five strains of E. coli.
|
|
Addition of SILAC Annotated Spectrum Libraries for X! Hunter (2009/04/02)
A new curation of the X! Hunter libraries now has a separate library file for annotated spectra that
are assigned to the heavy isotope labels in SILAC experiments. The new libraries have been made for
human, mouse and yeast peptides and they are available for download from the GPM ftp site eukaryote libraries collection.
The SILAC libraries are named human_silac_20.hlf, mouse_silac_20.hlf and yeast_silac_20.hlf.
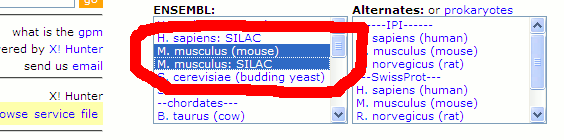
These libraries are also mounted on the public X! Hunter search site. To search a SILAC data set to extract both heavy and light
peptides, select both the normal and SILAC libraries, as illustrated below. To extract only the SILAC (or normal) data, use only
the appropriate one of these selections. In addition to the SILAC libraries, a major new release of the yeast library is also available.
|
|
Changes to Gene Ontology (GO) display (2009/04/22)
The display that indicates Gene Ontology classifications for the proteins in a data set has been updated to
include more GO categories. The original display used 25 GO categories, made up of a selection of
cellular components and cellular processes. This display has been updated to use 105 categories, with
individual displays for each of cellular components, cellular processes and molecular functions (35 categories each).
Once the GO page has been displayed (cellular components is the default), the other displays can be accessed using
a new set of links, just below the histogram at the top of the page:
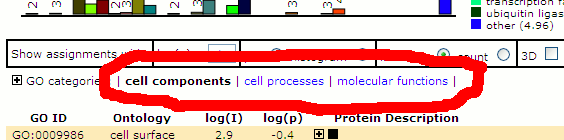
The new categories were selected based on the current population of GPMDB. Some of the GO descriptions have been
altered slightly, to improve legibility in the alphabetical order used in the displays. The current list of categories that
can be accessed with the new system are as follows:
| cellular components | cellular processes | molecular function |
- cell surface
- centrosome
- chromatin
- chromosome
- cytoplasm
- cytoskeleton
- cytoskeleton, actin
- endoplasmic reticulum
- endosome
- extracellular region
- extracellular matrix
- focal adhesion
- Golgi apparatus
- intermediate filament
- lysosome
- membrane
- membrane, anchored
- membrane, integral
- membrane, plasma
- membrane. nuclear
- microsome
- microtubule
- mitochondrion
- myosin complex
- nuclear pore
- nucleolus
- nucleus
- peroxisome
- proteasome
- ribonucleoprotein complex
- ribosome
- spliceosome
- tight junction
- transcription factor complex
- ubiquitin ligase complex
|
- apoptosis
- carbohydrate metabolism
- cell adhesion
- cell cycle
- cell differentiation
- cell-cell signaling
- cell proliferation, +ve regulation
- cell proliferation, -ve regulation
- chromatin modification
- dephosphorylation
- DNA repair
- DNA replication
- immune response
- inflammatory response
- integrin-mediated signaling
- lipid metabolic process
- meiosis
- metabolic process
- microtubule-based movement
- mitosis
- multicellular development
- protein dephosphorylation
- protein glycosylation
- protein phosphorylation
- protein folding
- proteolysis
- RNA splicing
- signal transduction
- signaling, G-protein
- transcription
- transcription, regulation
- translation
- transport
- transport, ion
- transport, protein
|
- acyltransferase activity
- binding, ATP
- binding, calcium ion
- binding, DNA
- binding, GTP
- binding, iron ion
- binding, magnesium ion
- binding, manganese ion
- binding, potassium ion
- binding, protein
- binding, RNA
- binding, sugar
- binding, zinc ion
- catalytic activity
- cytokine activity
- electron carrier activity
- G-protein coupled receptor activity
- hormone activity
- hydrolase activity
- ion channel activity
- kinase activity
- ligand-dependent nuclear receptor activity
- ligase activity
- lyase activity
- methyltransferase activity
- monooxygenase activity
- oxidoreductase activity
- peptidase activity
- phosphatase activity
- protein S/T kinase activity
- protein Y kinase activity
- receptor activity
- signal transducer activity
- transporter activity
- ubiquitin-protein ligase activity
|
|
|
Addition of SILAC Annotated Spectrum Libraries for X! Hunter (2009/04/02)
A new curation of the X! Hunter libraries now has a separate library file for annotated spectra that
are assigned to the heavy isotope labels in SILAC experiments. The new libraries have been made for
human, mouse and yeast peptides and they are available for download from the GPM ftp site eukaryote libraries collection.
The SILAC libraries are named human_silac_20.hlf, mouse_silac_20.hlf and yeast_silac_20.hlf.
These libraries are also mounted on the public X! Hunter search site. To search a SILAC data set to extract both heavy and light
peptides, select both the normal and SILAC libraries, as illustrated below. To extract only the SILAC (or normal) data, use only
the appropriate one of these selections. In addition to the SILAC libraries, a major new release of the yeast library is also available.

|
|
New Gene Ontology pages added to GPMDB
(2009/04/02)
GPMDB has had a limited set of Gene Ontology (GO) pages available, that contain
lists of observed proteins in the human, mouse or yeast proteomes that belong to
particular GO classifications.
The original index
has been maintained, but a large selection of new categories has been added. These new
new pages can be reached by clicking on the all human,
all mouse, or
all yeast links. These indexes
display all of the available GO classifications, broken up into biological process, cellular component and molecular function
sub-categories. The human and mouse pages use the full set of GO categories (from ENSEMBL), while the yeast
page uses GO-slim (from SGD).
|
|
Amino acid analysis (AAA) display added
(2009/03/24)
A new display that calculates eight amino acid analyses for a
particular data set has been made available in both GPMDB and all of
the public GPM search servers. The results of the analysis is displayed
in a table, giving the amino acid composition of the following sets of
residues found in a search model:
- Pre: AAA of the residue in the protein sequence immediately prior to the N-terminus of each unique peptide;
- N-terminal: AAA of the N-terminal residue of each unique peptide;
- C-terminal: AAA of the C-terminal residue of each unique peptide;
- Post: AAA of the residue in the protein sequence immediately following the C-terminus of each unique peptide;
- All: AAA of all peptides identified (including multiple identifications of the same peptide sequence);
- Protein: AAA of all proteins identified;
- Unique: AAA of the unique peptides identified; and
- Delta: difference between the unique peptide AAA and the protein AAA.
The display can be accessed through the "aaa" link on the
peptide display tool bar (click here
for an example).
|
|
New documentation resource for GPMDB
(2009/03/06)
In order to improve the documentation for GPM, we have started
a project on our wiki called Technical
Overview. Dan Evans will be adding new information and updating the
writeups for the GPM utilities and GPMDB table structure.
|
|
ENSEMBL sequences updated
(2009/01/30)
Protein sequences on the GPM search sites that use ENSEMBL
accession numbers have been updated to ENSEMBL version 52. The
associated sequence annotations have been updated to UNIPROT
version 14.7.
|
|
70,000,000th Peptide Id Recorded
(2009/01/14)
Today, GPMDB passed the 70 million mark for peptide
identifications. We would like to thank all of the data
contributors who have made this project a success. Special
thanks goes to our ProteomExchange partners TRANCHE and PRIDE,
for making their data available.
|
|
GPMDB Phosphopeptide Collection
(pSYT) (2009/01/14)
GPMDB has a large number of phophopeptide observations
available for use. We have added a new user interface, called
pSYT, to allow users direct access to this information on a
protein by protein basis. To access pSYT for human, mouse,
yeast and zebrafish proteins, use the corresponding link on the
protein toolbar at the top of any protein display page:
The current statistics for phosphopeptides in GPMDB are as
follows:
|
Species
|
Observations
|
Unique peptides
|
Observations/peptide
|
|
H. sapiens
|
382,884
|
17,876
|
21.4 ×
|
|
M. musculus
|
61,003
|
9,125
|
6.7 ×
|
|
S. cerevisiae
|
20,043
|
4536
|
4.4 ×
|
|
D. rerio
|
10,588
|
959
|
11.0 ×
|
|
|
Copyright © 2009, The Global Proteome Machine Organization
|
|

